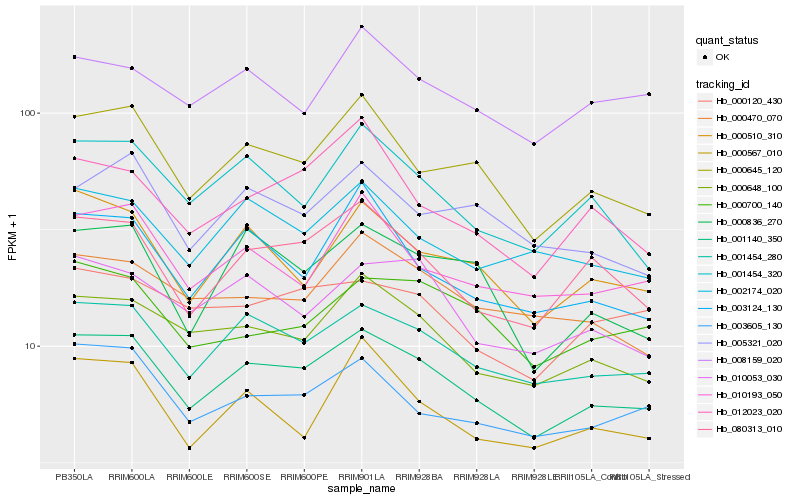
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001140_350 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648465 [Jatropha curcas] |
| 2 |
Hb_001454_280 |
0.0427917901 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 3 |
Hb_000645_120 |
0.0614423666 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 32 [Jatropha curcas] |
| 4 |
Hb_080313_010 |
0.0622428669 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 5 |
Hb_000648_100 |
0.0626357366 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 6 |
Hb_000510_310 |
0.0711425753 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 7 |
Hb_003605_130 |
0.0725255529 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Populus euphratica] |
| 8 |
Hb_010053_030 |
0.0742406114 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 9 |
Hb_002174_020 |
0.0750173684 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 10 |
Hb_001454_320 |
0.0751714865 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_010193_050 |
0.0765573567 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003124_130 |
0.0801608799 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000836_270 |
0.0805282404 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2 [Jatropha curcas] |
| 14 |
Hb_000470_070 |
0.0809178871 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |
| 15 |
Hb_000120_430 |
0.0816647438 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 16 |
Hb_008159_020 |
0.0816920236 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 17 |
Hb_000567_010 |
0.081722363 |
- |
- |
PREDICTED: uncharacterized protein LOC105631110 [Jatropha curcas] |
| 18 |
Hb_000700_140 |
0.0828065929 |
- |
- |
PREDICTED: guanine nucleotide-binding protein-like NSN1 [Jatropha curcas] |
| 19 |
Hb_012023_020 |
0.0828667925 |
- |
- |
hypothetical protein JCGZ_03076 [Jatropha curcas] |
| 20 |
Hb_005321_020 |
0.0828947198 |
- |
- |
splicing factor-related family protein [Populus trichocarpa] |