| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
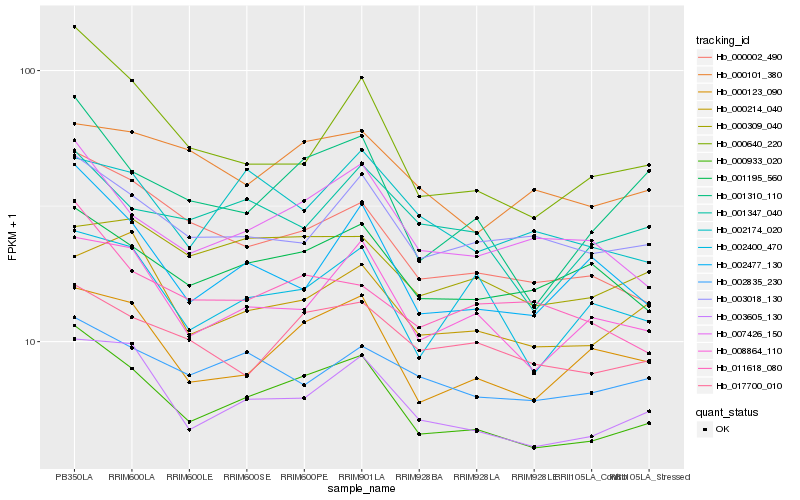
Hb_000933_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003605_130 |
0.0575421256 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Populus euphratica] |
| 3 |
Hb_000123_090 |
0.0718144893 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os03g0620400-like [Jatropha curcas] |
| 4 |
Hb_008864_110 |
0.0718344879 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 5 |
Hb_007426_150 |
0.0730904485 |
- |
- |
PREDICTED: F-box protein At5g46170-like [Jatropha curcas] |
| 6 |
Hb_000002_490 |
0.0738306849 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |
| 7 |
Hb_001195_560 |
0.0766495774 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003018_130 |
0.0792082376 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 9 |
Hb_001310_110 |
0.0842449863 |
- |
- |
unnamed protein product [Coffea canephora] |
| 10 |
Hb_017700_010 |
0.0848361863 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 11 |
Hb_000214_040 |
0.0860233589 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001347_040 |
0.0872591671 |
- |
- |
PREDICTED: uncharacterized protein LOC105633920 [Jatropha curcas] |
| 13 |
Hb_002400_470 |
0.0892073261 |
- |
- |
PREDICTED: calcium uptake protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 14 |
Hb_000309_040 |
0.0900544623 |
- |
- |
PREDICTED: protein farnesyltransferase subunit beta isoform X1 [Jatropha curcas] |
| 15 |
Hb_011618_080 |
0.0901808641 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 16 |
Hb_002835_230 |
0.0907657373 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_002477_130 |
0.0914237343 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000640_220 |
0.0915185449 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-4 [Jatropha curcas] |
| 19 |
Hb_002174_020 |
0.0918918287 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 20 |
Hb_000101_380 |
0.0920429319 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |