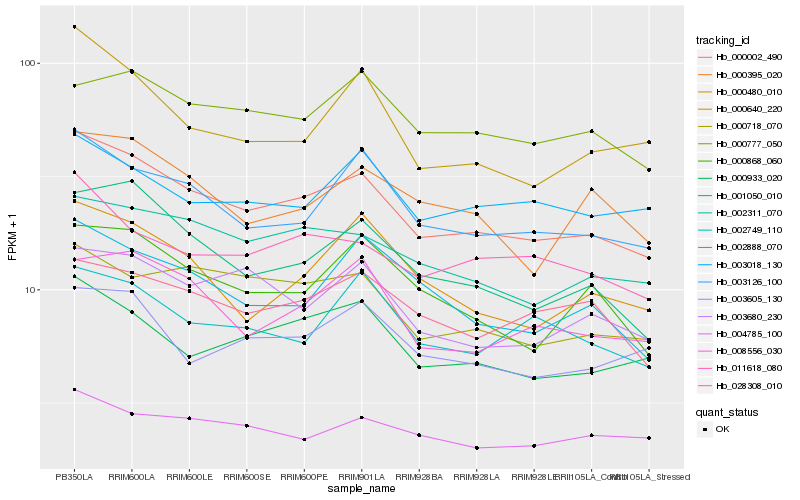
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000002_490 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |
| 2 |
Hb_003126_100 |
0.0640719885 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 3 |
Hb_002311_070 |
0.0683381814 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 4 |
Hb_001050_010 |
0.0730073272 |
- |
- |
Advillin [Gossypium arboreum] |
| 5 |
Hb_000933_020 |
0.0738306849 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000718_070 |
0.0762035772 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_011618_080 |
0.0780663019 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 8 |
Hb_008556_030 |
0.0788085671 |
- |
- |
PREDICTED: uncharacterized protein LOC105648021 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003680_230 |
0.0800027068 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 10 |
Hb_028308_010 |
0.0808284828 |
- |
- |
PREDICTED: probable protein S-acyltransferase 4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000868_060 |
0.0811257515 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 12 |
Hb_002888_070 |
0.0822017525 |
- |
- |
PREDICTED: tubulin-folding cofactor E isoform X1 [Pyrus x bretschneideri] |
| 13 |
Hb_000395_020 |
0.0836453652 |
- |
- |
PREDICTED: ankyrin repeat and KH domain-containing protein R11A8.7 [Jatropha curcas] |
| 14 |
Hb_003605_130 |
0.0842426092 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Populus euphratica] |
| 15 |
Hb_004785_100 |
0.0846691397 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002749_110 |
0.0854074164 |
- |
- |
PREDICTED: uncharacterized protein LOC105636001 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000640_220 |
0.0854634214 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-4 [Jatropha curcas] |
| 18 |
Hb_000777_050 |
0.0857970123 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |
| 19 |
Hb_000480_010 |
0.0863313795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003018_130 |
0.0868430272 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |