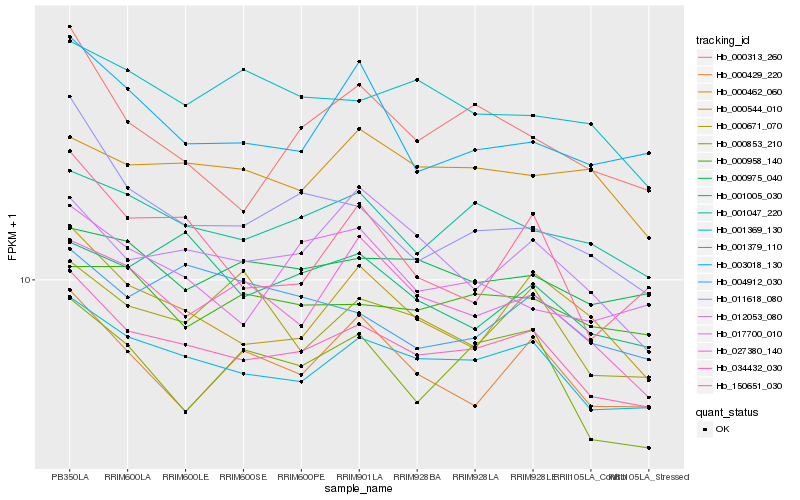
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034432_030 |
0.0 |
- |
- |
PREDICTED: probable glycosyltransferase At5g20260 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001379_110 |
0.0270969823 |
- |
- |
PREDICTED: sec1 family domain-containing protein MIP3 [Jatropha curcas] |
| 3 |
Hb_001047_220 |
0.0585141434 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 4 |
Hb_000544_010 |
0.0655662409 |
- |
- |
PREDICTED: GPI mannosyltransferase 1 [Jatropha curcas] |
| 5 |
Hb_011618_080 |
0.0702611141 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 6 |
Hb_000975_040 |
0.0715510499 |
- |
- |
hypothetical protein CICLE_v10028249mg [Citrus clementina] |
| 7 |
Hb_012053_080 |
0.0725435947 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 8 |
Hb_027380_140 |
0.0726854222 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000313_260 |
0.0740996925 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 10 |
Hb_001369_130 |
0.0741831486 |
- |
- |
PREDICTED: autophagy-related protein 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000429_220 |
0.074419376 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105640858 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003018_130 |
0.0750869646 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 13 |
Hb_000958_140 |
0.0756518448 |
- |
- |
PREDICTED: uncharacterized protein LOC105628714 [Jatropha curcas] |
| 14 |
Hb_000853_210 |
0.0764196194 |
- |
- |
PREDICTED: uncharacterized protein LOC105642575 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000671_070 |
0.076723754 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 16 |
Hb_017700_010 |
0.077428996 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 17 |
Hb_001005_030 |
0.0779141361 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 18 |
Hb_000462_060 |
0.0787079793 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 19 |
Hb_004912_030 |
0.079528907 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 20 |
Hb_150651_030 |
0.0796086837 |
- |
- |
PREDICTED: metallophosphoesterase 1-like [Jatropha curcas] |