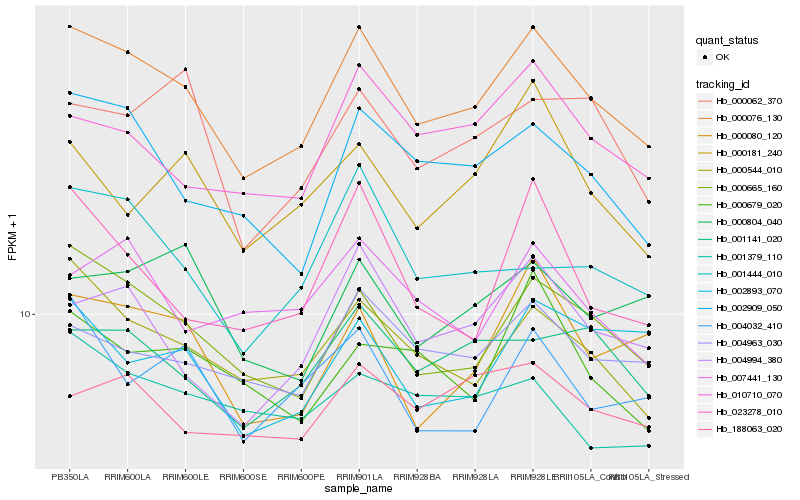
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000076_130 |
0.0 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 2 |
Hb_000665_160 |
0.054925188 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic isoform X1 [Jatropha curcas] |
| 3 |
Hb_000544_010 |
0.0629932715 |
- |
- |
PREDICTED: GPI mannosyltransferase 1 [Jatropha curcas] |
| 4 |
Hb_002909_050 |
0.0703132221 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004994_380 |
0.0733872931 |
- |
- |
hypothetical protein JCGZ_14410 [Jatropha curcas] |
| 6 |
Hb_004963_030 |
0.0756037895 |
- |
- |
PREDICTED: twinkle homolog protein, chloroplastic/mitochondrial [Jatropha curcas] |
| 7 |
Hb_001141_020 |
0.0765832113 |
- |
- |
PREDICTED: stress-induced-phosphoprotein 1 [Jatropha curcas] |
| 8 |
Hb_010710_070 |
0.0792608357 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |
| 9 |
Hb_001444_010 |
0.0814683589 |
transcription factor |
TF Family: DDT |
PREDICTED: DDT domain-containing protein DDB_G0282237 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000080_120 |
0.0824459853 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350-like [Jatropha curcas] |
| 11 |
Hb_023278_010 |
0.0849613137 |
- |
- |
Chaperone protein DnaJ [Glycine soja] |
| 12 |
Hb_004032_410 |
0.0850016165 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000062_370 |
0.0857947095 |
- |
- |
beta-ketoacyl-ACP reductase [Vernicia fordii] |
| 14 |
Hb_188063_020 |
0.0876291219 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007441_130 |
0.0879289572 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_000804_040 |
0.0886104425 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_002893_070 |
0.0891705577 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000679_020 |
0.0893679175 |
transcription factor |
TF Family: HB |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 19 |
Hb_000181_240 |
0.0902298663 |
- |
- |
PREDICTED: uncharacterized protein LOC105637672 [Jatropha curcas] |
| 20 |
Hb_001379_110 |
0.0907943827 |
- |
- |
PREDICTED: sec1 family domain-containing protein MIP3 [Jatropha curcas] |