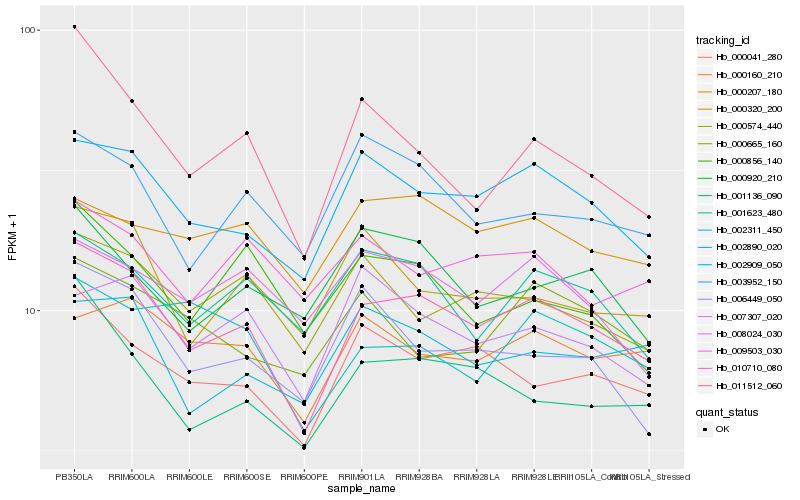
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010710_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_002909_050 |
0.0747962967 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000041_280 |
0.0879240994 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 6 [Jatropha curcas] |
| 4 |
Hb_001136_090 |
0.0971153864 |
- |
- |
Protein PNS1, putative [Ricinus communis] |
| 5 |
Hb_001623_480 |
0.0988338225 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 13 [Jatropha curcas] |
| 6 |
Hb_011512_060 |
0.101059097 |
- |
- |
PREDICTED: chaperonin CPN60-2, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000574_440 |
0.1013380185 |
- |
- |
PREDICTED: uncharacterized protein LOC105634833 [Jatropha curcas] |
| 8 |
Hb_000160_210 |
0.1029111724 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002890_020 |
0.1038257977 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 5 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000320_200 |
0.1040804438 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 6 homolog [Jatropha curcas] |
| 11 |
Hb_003952_150 |
0.104336019 |
- |
- |
PREDICTED: cactin [Jatropha curcas] |
| 12 |
Hb_000207_180 |
0.1050220439 |
- |
- |
PREDICTED: calponin homology domain-containing protein DDB_G0272472 [Jatropha curcas] |
| 13 |
Hb_006449_050 |
0.1051493849 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX2-like [Jatropha curcas] |
| 14 |
Hb_007307_020 |
0.1052835698 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_009503_030 |
0.1081626727 |
- |
- |
protein dimerization, putative [Ricinus communis] |
| 16 |
Hb_002311_450 |
0.1084024904 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 17 |
Hb_008024_030 |
0.1088393788 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105648348 [Jatropha curcas] |
| 18 |
Hb_000856_140 |
0.1103479751 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000920_210 |
0.1103850666 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 20 |
Hb_000665_160 |
0.1114016688 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic isoform X1 [Jatropha curcas] |