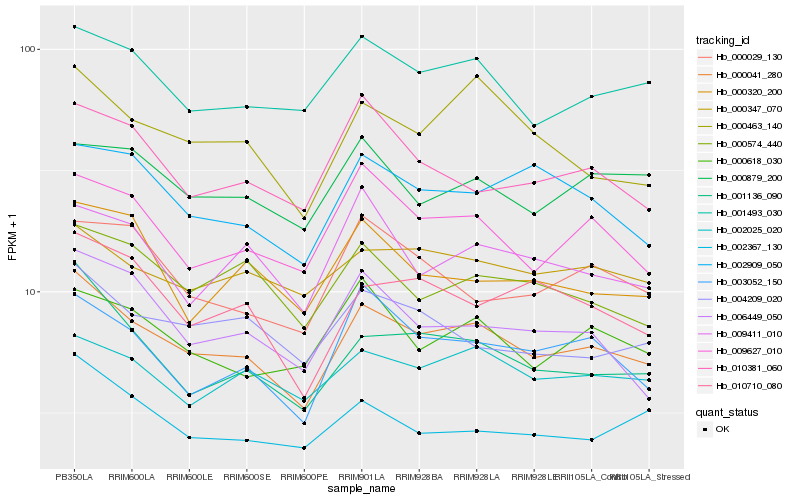
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000041_280 |
0.0 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 6 [Jatropha curcas] |
| 2 |
Hb_001136_090 |
0.0737912695 |
- |
- |
Protein PNS1, putative [Ricinus communis] |
| 3 |
Hb_001493_030 |
0.0788976973 |
- |
- |
hypothetical protein MIMGU_mgv1a005351mg [Erythranthe guttata] |
| 4 |
Hb_000463_140 |
0.0802532247 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 5 |
Hb_009627_010 |
0.0822509217 |
- |
- |
Protein dom-3, putative [Ricinus communis] |
| 6 |
Hb_003052_150 |
0.0831991789 |
- |
- |
PREDICTED: putative ATP-dependent RNA helicase DHX33 [Jatropha curcas] |
| 7 |
Hb_000574_440 |
0.0863829746 |
- |
- |
PREDICTED: uncharacterized protein LOC105634833 [Jatropha curcas] |
| 8 |
Hb_004209_020 |
0.0870462801 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 51-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_002025_020 |
0.0874418479 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39710 [Jatropha curcas] |
| 10 |
Hb_010710_080 |
0.0879240994 |
- |
- |
- |
| 11 |
Hb_000618_030 |
0.0886208439 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOUSLED-like isoform X4 [Populus euphratica] |
| 12 |
Hb_002909_050 |
0.0890206129 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006449_050 |
0.0895973893 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX2-like [Jatropha curcas] |
| 14 |
Hb_000347_070 |
0.0908706241 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_010381_060 |
0.0921027294 |
- |
- |
DEAD-box ATP-dependent RNA helicase 20 -like protein [Gossypium arboreum] |
| 16 |
Hb_002367_130 |
0.0935200579 |
- |
- |
UNC-50 family protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_000879_200 |
0.0948035228 |
- |
- |
PREDICTED: A-kinase anchor protein 17A [Jatropha curcas] |
| 18 |
Hb_000029_130 |
0.0955786014 |
- |
- |
PREDICTED: ruvB-like protein 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_009411_010 |
0.0962688534 |
- |
- |
PREDICTED: uncharacterized protein LOC105645272 [Jatropha curcas] |
| 20 |
Hb_000320_200 |
0.0962770872 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 6 homolog [Jatropha curcas] |