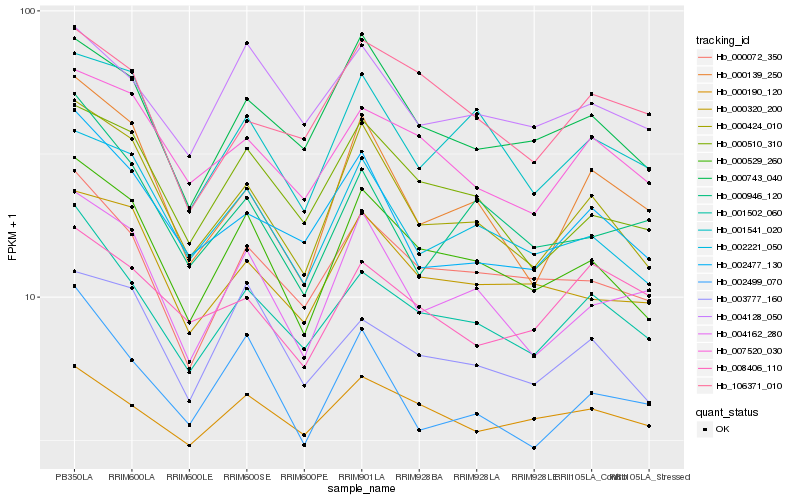
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001502_060 |
0.0 |
- |
- |
DNA mismatch repair protein MSH2, putative [Ricinus communis] |
| 2 |
Hb_000072_350 |
0.0617111303 |
- |
- |
PREDICTED: WPP domain-interacting tail-anchored protein 1 [Jatropha curcas] |
| 3 |
Hb_007520_030 |
0.0713858933 |
- |
- |
PREDICTED: peroxisomal membrane protein PEX14 [Jatropha curcas] |
| 4 |
Hb_000529_260 |
0.0756813533 |
- |
- |
PREDICTED: protein SGT1 homolog At5g65490 [Jatropha curcas] |
| 5 |
Hb_002477_130 |
0.0762462161 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000424_010 |
0.0822552194 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004128_050 |
0.0822834149 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_000743_040 |
0.0843534874 |
- |
- |
ribosomal RNA methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000510_310 |
0.0860912028 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 10 |
Hb_106371_010 |
0.0882545786 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 11 |
Hb_001541_020 |
0.089335757 |
- |
- |
PREDICTED: uncharacterized protein At3g49055 [Jatropha curcas] |
| 12 |
Hb_002221_050 |
0.0895678323 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP65 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002499_070 |
0.089791076 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000320_200 |
0.0901432069 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 6 homolog [Jatropha curcas] |
| 15 |
Hb_003777_160 |
0.0903302789 |
transcription factor |
TF Family: bZIP |
PREDICTED: G-box-binding factor 4 isoform X1 [Jatropha curcas] |
| 16 |
Hb_008406_110 |
0.0909699835 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a isoform X1 [Jatropha curcas] |
| 17 |
Hb_000946_120 |
0.0918317259 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_004162_280 |
0.0920355179 |
- |
- |
PREDICTED: uncharacterized protein LOC105628004 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000139_250 |
0.0934595613 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |
| 20 |
Hb_000190_120 |
0.0935892116 |
- |
- |
PREDICTED: uncharacterized protein LOC105649936 [Jatropha curcas] |