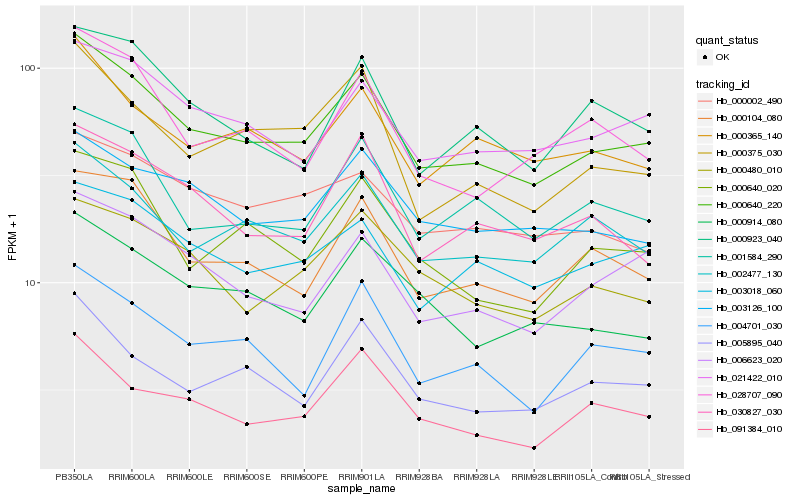
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000640_220 |
0.0 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-4 [Jatropha curcas] |
| 2 |
Hb_021422_010 |
0.0703213517 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 1, mitochondrial [Jatropha curcas] |
| 3 |
Hb_002477_130 |
0.0720337601 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000104_080 |
0.0733144137 |
- |
- |
PREDICTED: WD repeat domain-containing protein 83 [Jatropha curcas] |
| 5 |
Hb_001584_290 |
0.0742903841 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 6 |
Hb_000365_140 |
0.0765489415 |
- |
- |
PREDICTED: ethanolamine-phosphate cytidylyltransferase [Jatropha curcas] |
| 7 |
Hb_003126_100 |
0.0781403308 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 8 |
Hb_003018_060 |
0.0785329956 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase A5 [Jatropha curcas] |
| 9 |
Hb_006623_020 |
0.0786399653 |
- |
- |
PREDICTED: protein LURP-one-related 7 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005895_040 |
0.0802085368 |
- |
- |
PREDICTED: tRNA threonylcarbamoyladenosine dehydratase [Jatropha curcas] |
| 11 |
Hb_000640_020 |
0.0814317749 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 12 |
Hb_091384_010 |
0.0816924734 |
- |
- |
hypothetical protein JCGZ_09207 [Jatropha curcas] |
| 13 |
Hb_000375_030 |
0.0822994558 |
- |
- |
- |
| 14 |
Hb_000914_080 |
0.0833406149 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_000480_010 |
0.0835669739 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_028707_090 |
0.0835857483 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Jatropha curcas] |
| 17 |
Hb_000923_040 |
0.0842819545 |
- |
- |
PREDICTED: 15 kDa selenoprotein [Jatropha curcas] |
| 18 |
Hb_004701_030 |
0.0849080418 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 19 |
Hb_000002_490 |
0.0854634214 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |
| 20 |
Hb_030827_030 |
0.0856664483 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |