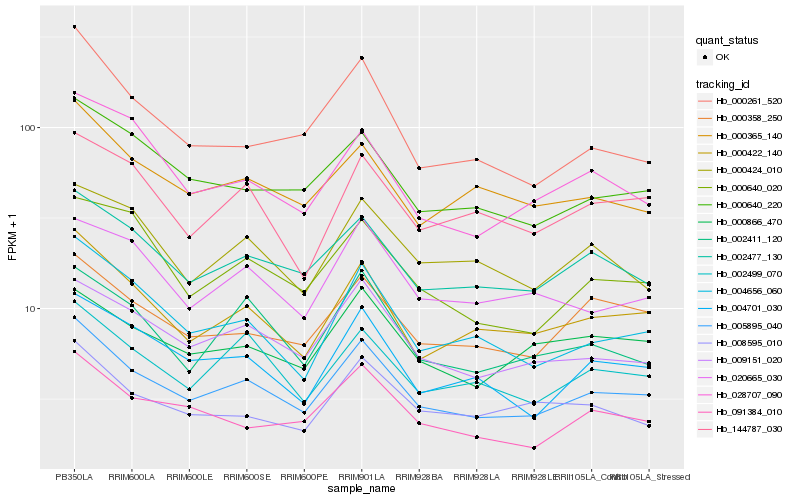
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005895_040 |
0.0 |
- |
- |
PREDICTED: tRNA threonylcarbamoyladenosine dehydratase [Jatropha curcas] |
| 2 |
Hb_000422_140 |
0.0629782851 |
- |
- |
PREDICTED: apoptosis-inducing factor 2-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_009151_020 |
0.0697025902 |
- |
- |
PREDICTED: DNA polymerase delta catalytic subunit [Jatropha curcas] |
| 4 |
Hb_002477_130 |
0.0697904205 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004656_060 |
0.0741111311 |
- |
- |
PREDICTED: uncharacterized protein LOC105634581 [Jatropha curcas] |
| 6 |
Hb_004701_030 |
0.077207433 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 7 |
Hb_002499_070 |
0.0783522329 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000640_220 |
0.0802085368 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-4 [Jatropha curcas] |
| 9 |
Hb_091384_010 |
0.0823200279 |
- |
- |
hypothetical protein JCGZ_09207 [Jatropha curcas] |
| 10 |
Hb_000365_140 |
0.0837193956 |
- |
- |
PREDICTED: ethanolamine-phosphate cytidylyltransferase [Jatropha curcas] |
| 11 |
Hb_000358_250 |
0.0845411094 |
- |
- |
PREDICTED: cell division cycle protein 27 homolog B [Jatropha curcas] |
| 12 |
Hb_002411_120 |
0.086653147 |
- |
- |
PREDICTED: uncharacterized protein LOC105631645 [Jatropha curcas] |
| 13 |
Hb_000261_520 |
0.0894277874 |
- |
- |
PREDICTED: uncharacterized protein LOC105633192 [Jatropha curcas] |
| 14 |
Hb_020665_030 |
0.0896632582 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000640_020 |
0.0899921356 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 16 |
Hb_028707_090 |
0.0914002217 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Jatropha curcas] |
| 17 |
Hb_000866_470 |
0.0914613757 |
- |
- |
PREDICTED: uncharacterized protein LOC105643015 [Jatropha curcas] |
| 18 |
Hb_144787_030 |
0.0923080046 |
- |
- |
PREDICTED: protein SLOW GREEN 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000424_010 |
0.0924694553 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 20 |
Hb_008595_010 |
0.092788558 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g06710, mitochondrial [Jatropha curcas] |