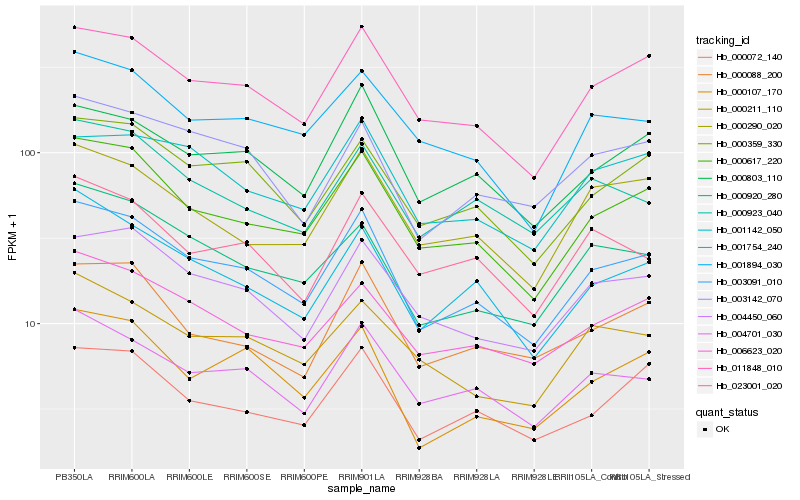
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003091_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 2 |
Hb_000617_220 |
0.0517206629 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 3 |
Hb_011848_010 |
0.055497437 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 4 |
Hb_004701_030 |
0.0659248146 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 5 |
Hb_000359_330 |
0.0693284501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004450_060 |
0.0744685083 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001754_240 |
0.0745661041 |
- |
- |
PREDICTED: mitochondrial thiamine pyrophosphate carrier isoform X1 [Jatropha curcas] |
| 8 |
Hb_006623_020 |
0.0750703797 |
- |
- |
PREDICTED: protein LURP-one-related 7 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000803_110 |
0.0762944537 |
- |
- |
PREDICTED: RNA-binding protein 8A [Jatropha curcas] |
| 10 |
Hb_000920_280 |
0.0778361811 |
- |
- |
Protein SENSITIVITY TO RED LIGHT REDUCED, putative [Ricinus communis] |
| 11 |
Hb_001894_030 |
0.0797600845 |
- |
- |
Nucleic acid-binding, OB-fold-like protein [Theobroma cacao] |
| 12 |
Hb_003142_070 |
0.0828760503 |
- |
- |
hypothetical protein F383_14657 [Gossypium arboreum] |
| 13 |
Hb_000072_140 |
0.0829843176 |
- |
- |
- |
| 14 |
Hb_000290_020 |
0.083302001 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit K-like isoform X2 [Populus euphratica] |
| 15 |
Hb_000088_200 |
0.0856614903 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_000107_170 |
0.0856804012 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC25 homolog [Jatropha curcas] |
| 17 |
Hb_001142_050 |
0.0862155374 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 18 |
Hb_000211_110 |
0.0874241588 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 35 kDa protein isoform X1 [Jatropha curcas] |
| 19 |
Hb_000923_040 |
0.0887412984 |
- |
- |
PREDICTED: 15 kDa selenoprotein [Jatropha curcas] |
| 20 |
Hb_023001_020 |
0.0889781284 |
- |
- |
PREDICTED: mitochondrial succinate-fumarate transporter 1 [Jatropha curcas] |