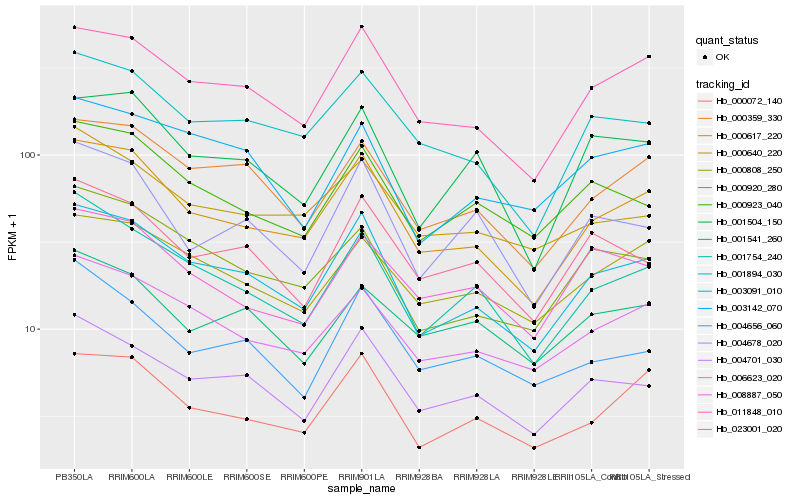
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001754_240 |
0.0 |
- |
- |
PREDICTED: mitochondrial thiamine pyrophosphate carrier isoform X1 [Jatropha curcas] |
| 2 |
Hb_003091_010 |
0.0745661041 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 3 |
Hb_006623_020 |
0.0767958397 |
- |
- |
PREDICTED: protein LURP-one-related 7 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004701_030 |
0.0788688633 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 5 |
Hb_000617_220 |
0.0799949331 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 6 |
Hb_000923_040 |
0.0880910171 |
- |
- |
PREDICTED: 15 kDa selenoprotein [Jatropha curcas] |
| 7 |
Hb_004656_060 |
0.0912059388 |
- |
- |
PREDICTED: uncharacterized protein LOC105634581 [Jatropha curcas] |
| 8 |
Hb_000920_280 |
0.0934057799 |
- |
- |
Protein SENSITIVITY TO RED LIGHT REDUCED, putative [Ricinus communis] |
| 9 |
Hb_000359_330 |
0.0936076341 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004678_020 |
0.0962100172 |
- |
- |
PREDICTED: protein YIF1B [Jatropha curcas] |
| 11 |
Hb_000640_220 |
0.0972619186 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-4 [Jatropha curcas] |
| 12 |
Hb_023001_020 |
0.0990631839 |
- |
- |
PREDICTED: mitochondrial succinate-fumarate transporter 1 [Jatropha curcas] |
| 13 |
Hb_000808_250 |
0.1024414894 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 14 |
Hb_001504_150 |
0.1048672413 |
- |
- |
PREDICTED: DCN1-like protein 4 [Jatropha curcas] |
| 15 |
Hb_011848_010 |
0.1051898444 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 16 |
Hb_003142_070 |
0.1053506798 |
- |
- |
hypothetical protein F383_14657 [Gossypium arboreum] |
| 17 |
Hb_001894_030 |
0.1055970185 |
- |
- |
Nucleic acid-binding, OB-fold-like protein [Theobroma cacao] |
| 18 |
Hb_008887_050 |
0.1059410448 |
- |
- |
PREDICTED: DNA-binding protein BIN4 [Jatropha curcas] |
| 19 |
Hb_000072_140 |
0.1065063981 |
- |
- |
- |
| 20 |
Hb_001541_260 |
0.1072124374 |
- |
- |
PREDICTED: protein MAK16 homolog [Jatropha curcas] |