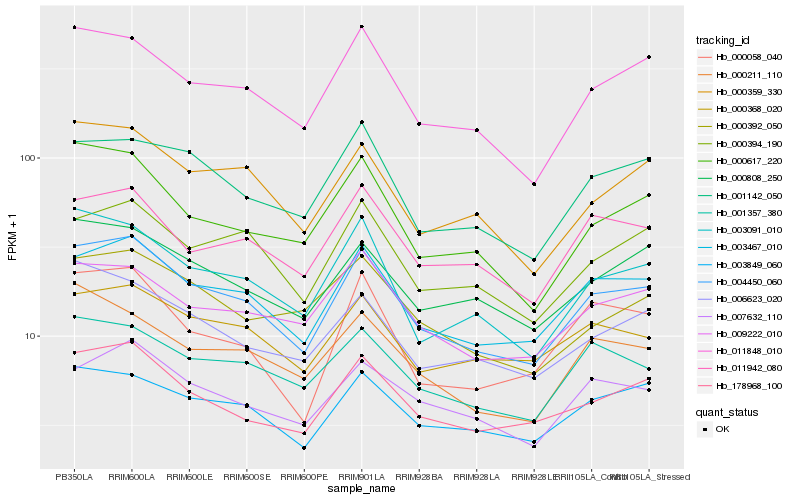
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004450_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 2 |
Hb_003467_010 |
0.0479485303 |
- |
- |
PREDICTED: probable thiol methyltransferase 2 [Jatropha curcas] |
| 3 |
Hb_011848_010 |
0.0661408758 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 4 |
Hb_178968_100 |
0.0677712205 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase isoform X1 [Populus euphratica] |
| 5 |
Hb_003091_010 |
0.0744685083 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 6 |
Hb_003849_060 |
0.0764446042 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000359_330 |
0.076982083 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000808_250 |
0.0791224014 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 9 |
Hb_007632_110 |
0.0794116696 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 10 |
Hb_001357_380 |
0.0794153954 |
- |
- |
PREDICTED: probable tetraacyldisaccharide 4'-kinase, mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_001142_050 |
0.0797559993 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 12 |
Hb_000394_190 |
0.0799844046 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_000617_220 |
0.0802194938 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 14 |
Hb_000368_020 |
0.0823972521 |
- |
- |
uroporphyrin-III methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_009222_010 |
0.0831695787 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 16 |
Hb_006623_020 |
0.0837050326 |
- |
- |
PREDICTED: protein LURP-one-related 7 isoform X1 [Jatropha curcas] |
| 17 |
Hb_011942_080 |
0.0841394153 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 18 |
Hb_000211_110 |
0.0866616168 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 35 kDa protein isoform X1 [Jatropha curcas] |
| 19 |
Hb_000392_050 |
0.0884784631 |
- |
- |
PREDICTED: uncharacterized protein LOC105640540 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000058_040 |
0.0887547111 |
- |
- |
PREDICTED: probable lipid-A-disaccharide synthase, mitochondrial [Jatropha curcas] |