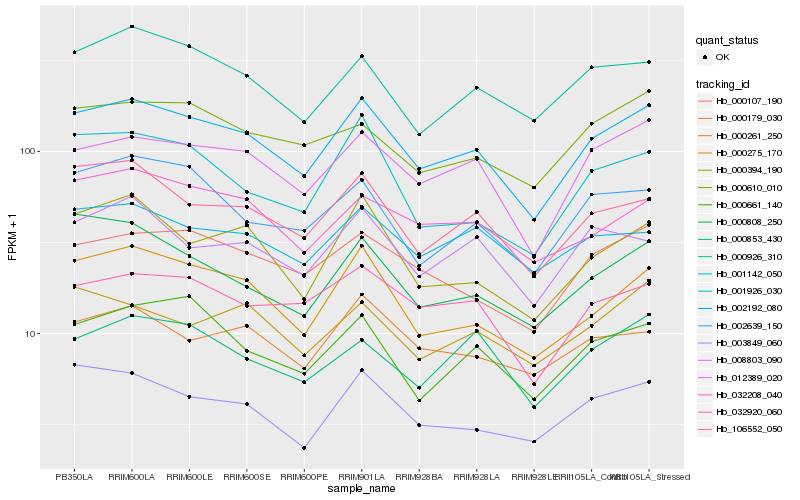
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002192_080 |
0.0 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 2 |
Hb_012389_020 |
0.0579304775 |
- |
- |
hypothetical protein POPTR_0006s05190g [Populus trichocarpa] |
| 3 |
Hb_000261_250 |
0.0652085271 |
- |
- |
elongation factor 1-alpha [Lepidium sativum] |
| 4 |
Hb_106552_050 |
0.069791854 |
- |
- |
PREDICTED: uncharacterized protein LOC105642134 [Jatropha curcas] |
| 5 |
Hb_000107_190 |
0.071448142 |
- |
- |
50S ribosomal protein L20 [Hevea brasiliensis] |
| 6 |
Hb_003849_060 |
0.0744824045 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 7 |
Hb_032920_060 |
0.0751846134 |
- |
- |
PREDICTED: mitotic checkpoint protein BUB3.1 [Jatropha curcas] |
| 8 |
Hb_000394_190 |
0.076004709 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_000926_310 |
0.0787794132 |
- |
- |
uncharacterized protein LOC100500241 [Glycine max] |
| 10 |
Hb_032208_040 |
0.0810728876 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL33 [Populus euphratica] |
| 11 |
Hb_002639_150 |
0.0819442122 |
- |
- |
- |
| 12 |
Hb_000610_010 |
0.0821225978 |
- |
- |
PREDICTED: SKP1-like protein 1B [Musa acuminata subsp. malaccensis] |
| 13 |
Hb_000661_140 |
0.0829561378 |
- |
- |
PREDICTED: uncharacterized protein LOC105648317 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000179_030 |
0.0830703755 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_008803_090 |
0.0835963617 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 6a [Jatropha curcas] |
| 16 |
Hb_001926_030 |
0.0840287723 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |
| 17 |
Hb_001142_050 |
0.0852822941 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 18 |
Hb_000808_250 |
0.0856282544 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 19 |
Hb_000853_430 |
0.0858378021 |
- |
- |
PREDICTED: uncharacterized protein LOC105642597 [Jatropha curcas] |
| 20 |
Hb_000275_170 |
0.0873225645 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |