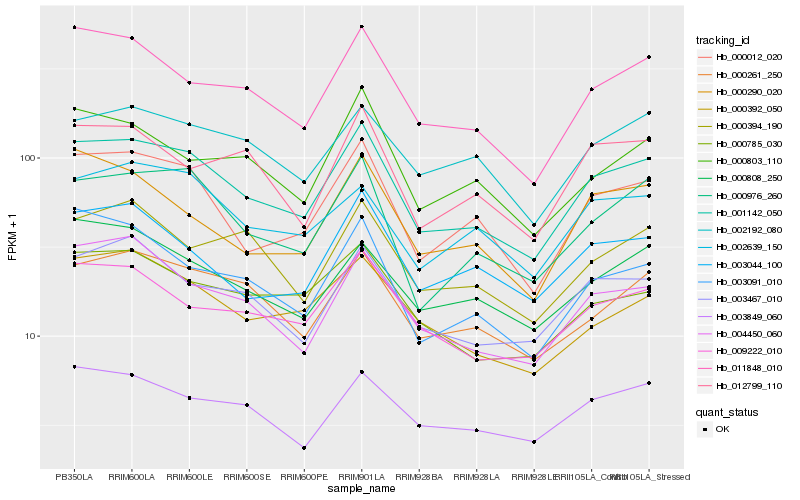
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001142_050 |
0.0 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 2 |
Hb_000012_020 |
0.0656389253 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 3 |
Hb_000976_260 |
0.0683904264 |
- |
- |
- |
| 4 |
Hb_011848_010 |
0.0706557926 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 5 |
Hb_000261_250 |
0.0734808557 |
- |
- |
elongation factor 1-alpha [Lepidium sativum] |
| 6 |
Hb_009222_010 |
0.0763909789 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 7 |
Hb_003467_010 |
0.0778456188 |
- |
- |
PREDICTED: probable thiol methyltransferase 2 [Jatropha curcas] |
| 8 |
Hb_004450_060 |
0.0797559993 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 9 |
Hb_003849_060 |
0.081255435 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 10 |
Hb_003044_100 |
0.0846976593 |
- |
- |
hypothetical protein F383_08446 [Gossypium arboreum] |
| 11 |
Hb_002192_080 |
0.0852822941 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 12 |
Hb_000808_250 |
0.0855667993 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 13 |
Hb_003091_010 |
0.0862155374 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 14 |
Hb_000785_030 |
0.0864156599 |
- |
- |
hypothetical protein RCOM_0808030 [Ricinus communis] |
| 15 |
Hb_000392_050 |
0.0869423563 |
- |
- |
PREDICTED: uncharacterized protein LOC105640540 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000803_110 |
0.0872769707 |
- |
- |
PREDICTED: RNA-binding protein 8A [Jatropha curcas] |
| 17 |
Hb_002639_150 |
0.0888374822 |
- |
- |
- |
| 18 |
Hb_000290_020 |
0.0897582653 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit K-like isoform X2 [Populus euphratica] |
| 19 |
Hb_000394_190 |
0.0898094164 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_012799_110 |
0.0903921777 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |