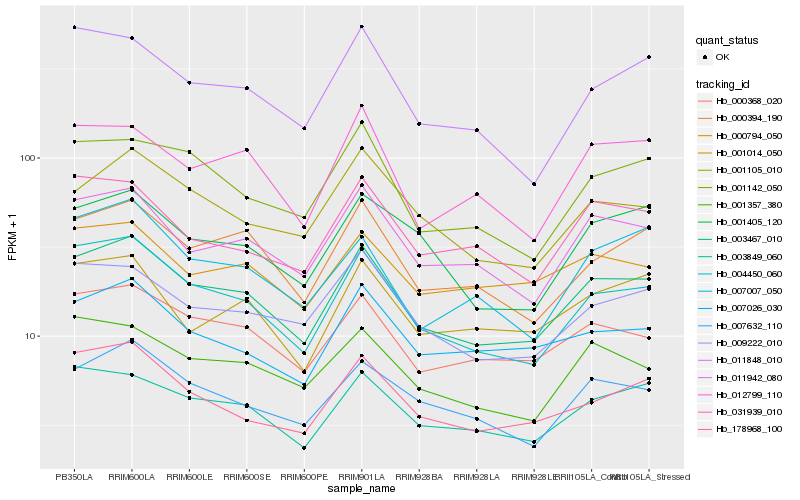
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003467_010 |
0.0 |
- |
- |
PREDICTED: probable thiol methyltransferase 2 [Jatropha curcas] |
| 2 |
Hb_004450_060 |
0.0479485303 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000368_020 |
0.063183098 |
- |
- |
uroporphyrin-III methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_011942_080 |
0.0632356789 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 5 |
Hb_003849_060 |
0.0668436193 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 6 |
Hb_007632_110 |
0.0684022485 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 7 |
Hb_000394_190 |
0.0701125221 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_178968_100 |
0.0733458774 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase isoform X1 [Populus euphratica] |
| 9 |
Hb_009222_010 |
0.0737869248 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 10 |
Hb_001357_380 |
0.0756736235 |
- |
- |
PREDICTED: probable tetraacyldisaccharide 4'-kinase, mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_001142_050 |
0.0778456188 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 12 |
Hb_001105_010 |
0.0792106497 |
desease resistance |
Gene Name: AIG1 |
PREDICTED: translocase of chloroplast 33, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_007026_030 |
0.0799186708 |
- |
- |
rubisco subunit binding-protein alpha subunit, ruba, putative [Ricinus communis] |
| 14 |
Hb_001405_120 |
0.0818949657 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit H [Jatropha curcas] |
| 15 |
Hb_012799_110 |
0.0819258298 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |
| 16 |
Hb_031939_010 |
0.0835282334 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 17 |
Hb_011848_010 |
0.0838260221 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 18 |
Hb_000794_050 |
0.0848175555 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 12 [Jatropha curcas] |
| 19 |
Hb_001014_050 |
0.0864267917 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105647306 [Jatropha curcas] |
| 20 |
Hb_007007_050 |
0.087667053 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Malus domestica] |