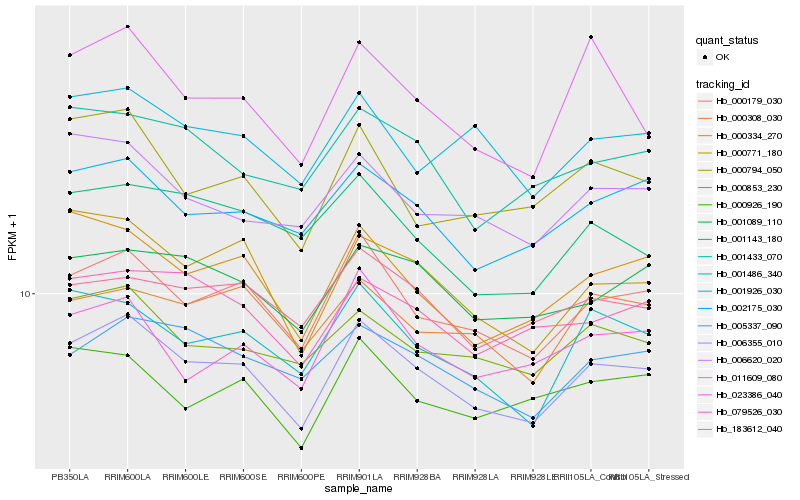
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006355_010 |
0.0 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |
| 2 |
Hb_000179_030 |
0.0446629396 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002175_030 |
0.06532327 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X2 [Jatropha curcas] |
| 4 |
Hb_000334_270 |
0.0657128303 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001486_340 |
0.066731699 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 1-like [Jatropha curcas] |
| 6 |
Hb_000771_180 |
0.0669910033 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 7 |
Hb_005337_090 |
0.06880387 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOUSLED-like [Jatropha curcas] |
| 8 |
Hb_000853_230 |
0.0688069708 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 9 |
Hb_001089_110 |
0.0691397105 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_079526_030 |
0.0705611121 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 11 |
Hb_001433_070 |
0.0729469121 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 12 |
Hb_000794_050 |
0.0739601865 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 12 [Jatropha curcas] |
| 13 |
Hb_000926_190 |
0.0741006205 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 3, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 14 |
Hb_006620_020 |
0.0751214034 |
- |
- |
PREDICTED: uncharacterized protein LOC105638878 [Jatropha curcas] |
| 15 |
Hb_000308_030 |
0.0752024839 |
- |
- |
PREDICTED: uncharacterized protein LOC105639686 [Jatropha curcas] |
| 16 |
Hb_183612_040 |
0.0752684656 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 17 |
Hb_011609_080 |
0.0758892287 |
- |
- |
uv excision repair protein rad23, putative [Ricinus communis] |
| 18 |
Hb_023386_040 |
0.075949548 |
- |
- |
PREDICTED: chaperone protein dnaJ 13-like [Populus euphratica] |
| 19 |
Hb_001926_030 |
0.0763618875 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |
| 20 |
Hb_001143_180 |
0.0774338293 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |