| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
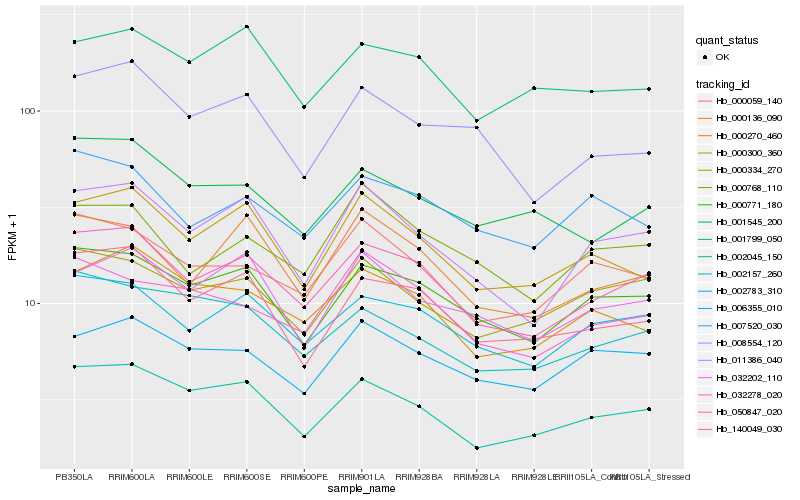
Hb_050847_020 |
0.0 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 2 |
Hb_002783_310 |
0.0520623108 |
- |
- |
PREDICTED: tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit trm6 [Jatropha curcas] |
| 3 |
Hb_000771_180 |
0.0605572101 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 4 |
Hb_002045_150 |
0.0660723688 |
- |
- |
histone acetyltransferase gcn5, putative [Ricinus communis] |
| 5 |
Hb_011386_040 |
0.0665536919 |
- |
- |
PREDICTED: uncharacterized protein LOC105639243 isoform X2 [Jatropha curcas] |
| 6 |
Hb_140049_030 |
0.0710432847 |
- |
- |
non-specific lipid-transfer protein-like protein At2g13820 precursor [Jatropha curcas] |
| 7 |
Hb_000136_090 |
0.0758157855 |
- |
- |
programmed cell death protein, putative [Ricinus communis] |
| 8 |
Hb_000059_140 |
0.079170784 |
- |
- |
rnase h, putative [Ricinus communis] |
| 9 |
Hb_000334_270 |
0.081155535 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000270_460 |
0.0812960683 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 8 [Jatropha curcas] |
| 11 |
Hb_000300_360 |
0.0821758919 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 12 |
Hb_006355_010 |
0.0857949324 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |
| 13 |
Hb_007520_030 |
0.0878698558 |
- |
- |
PREDICTED: peroxisomal membrane protein PEX14 [Jatropha curcas] |
| 14 |
Hb_000768_110 |
0.0879421116 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001545_200 |
0.0879532611 |
- |
- |
PREDICTED: uncharacterized oxidoreductase At4g09670 [Jatropha curcas] |
| 16 |
Hb_032278_020 |
0.0880878864 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 16 isoform X1 [Jatropha curcas] |
| 17 |
Hb_032202_110 |
0.0883081676 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 18 |
Hb_001799_050 |
0.0885262311 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 19 |
Hb_008554_120 |
0.0894632824 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 20 |
Hb_002157_260 |
0.0900598447 |
- |
- |
PREDICTED: partner of Y14 and mago-like [Jatropha curcas] |