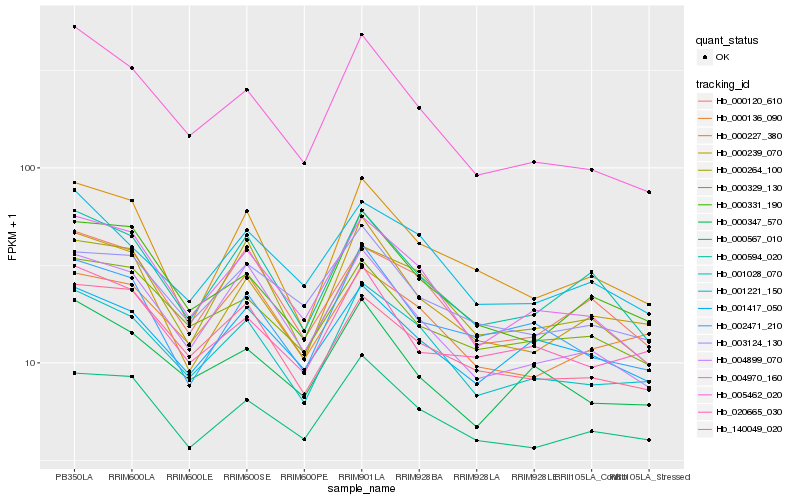
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001028_070 |
0.0 |
- |
- |
PREDICTED: WD repeat-containing protein 75 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001221_150 |
0.0691044356 |
- |
- |
AMP-activated protein kinase, gamma regulatory subunit, putative [Ricinus communis] |
| 3 |
Hb_000567_010 |
0.0693851675 |
- |
- |
PREDICTED: uncharacterized protein LOC105631110 [Jatropha curcas] |
| 4 |
Hb_004970_160 |
0.0697840684 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000136_090 |
0.0728053469 |
- |
- |
programmed cell death protein, putative [Ricinus communis] |
| 6 |
Hb_000120_610 |
0.0785720467 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 7 |
Hb_001417_050 |
0.0787480801 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 8 |
Hb_000331_190 |
0.0806500992 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 9 |
Hb_140049_020 |
0.0816088846 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 10 |
Hb_000594_020 |
0.0824412802 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004899_070 |
0.0843128813 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 12 |
Hb_000329_130 |
0.0845003028 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 13 |
Hb_000347_570 |
0.0846388617 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12b isoform X1 [Jatropha curcas] |
| 14 |
Hb_000264_100 |
0.0851536092 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000239_070 |
0.085548123 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000227_380 |
0.0858859661 |
- |
- |
PREDICTED: protein TIME FOR COFFEE isoform X3 [Jatropha curcas] |
| 17 |
Hb_005462_020 |
0.086071223 |
- |
- |
PREDICTED: UBP1-associated protein 2A-like [Jatropha curcas] |
| 18 |
Hb_003124_130 |
0.0864450521 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002471_210 |
0.0866618936 |
- |
- |
PREDICTED: THO complex subunit 2 [Jatropha curcas] |
| 20 |
Hb_020665_030 |
0.0871458422 |
- |
- |
conserved hypothetical protein [Ricinus communis] |