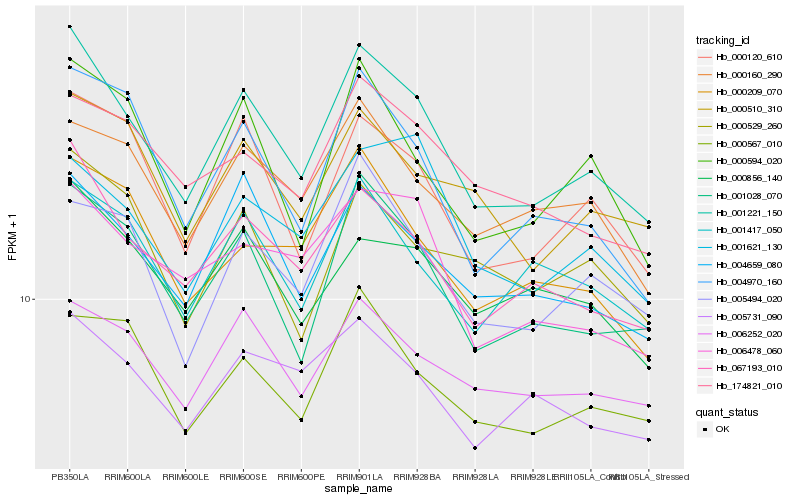
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001221_150 |
0.0 |
- |
- |
AMP-activated protein kinase, gamma regulatory subunit, putative [Ricinus communis] |
| 2 |
Hb_067193_010 |
0.0635539888 |
- |
- |
- |
| 3 |
Hb_001028_070 |
0.0691044356 |
- |
- |
PREDICTED: WD repeat-containing protein 75 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000120_610 |
0.0757608441 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 5 |
Hb_004659_080 |
0.0764639282 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 6 |
Hb_000594_020 |
0.0841332645 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001417_050 |
0.0843315129 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 8 |
Hb_006252_020 |
0.0843555542 |
- |
- |
PREDICTED: SET and MYND domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 9 |
Hb_004970_160 |
0.0847775416 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_000160_290 |
0.087974931 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_005494_020 |
0.0880910915 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 12 |
Hb_000567_010 |
0.0882311583 |
- |
- |
PREDICTED: uncharacterized protein LOC105631110 [Jatropha curcas] |
| 13 |
Hb_006478_060 |
0.0886494057 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005731_090 |
0.0888828804 |
- |
- |
PREDICTED: uncharacterized protein LOC105630164 [Jatropha curcas] |
| 15 |
Hb_000856_140 |
0.0889446139 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000510_310 |
0.0890880259 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 17 |
Hb_000209_070 |
0.0918267049 |
- |
- |
PREDICTED: chloride channel protein CLC-f isoform X1 [Jatropha curcas] |
| 18 |
Hb_000529_260 |
0.0935058199 |
- |
- |
PREDICTED: protein SGT1 homolog At5g65490 [Jatropha curcas] |
| 19 |
Hb_174821_010 |
0.0935650122 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 14 [Jatropha curcas] |
| 20 |
Hb_001621_130 |
0.0938872216 |
- |
- |
PREDICTED: pumilio homolog 1-like isoform X1 [Jatropha curcas] |