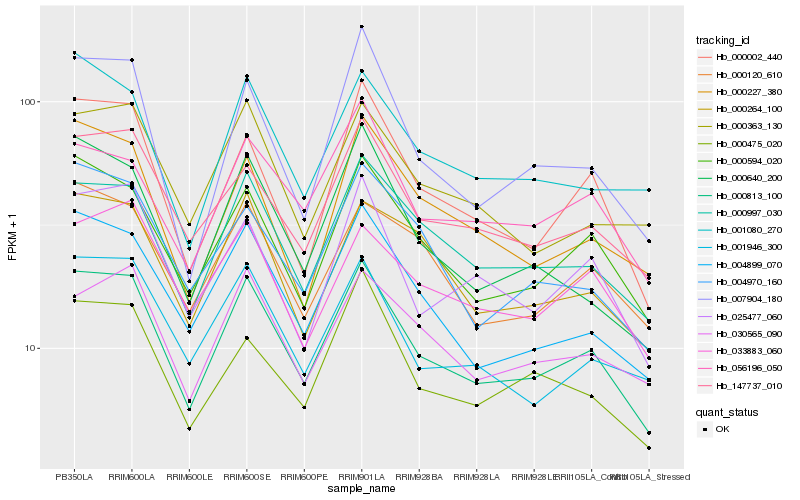
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000813_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000594_020 |
0.0579803179 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000997_030 |
0.0617956016 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 4 |
Hb_004899_070 |
0.0643366814 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 5 |
Hb_147737_010 |
0.0652627453 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000002_440 |
0.0720416733 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 7 |
Hb_001946_300 |
0.0752156367 |
- |
- |
PREDICTED: F-box protein At-B [Jatropha curcas] |
| 8 |
Hb_030565_090 |
0.0764363866 |
- |
- |
PREDICTED: DNA repair protein RAD50 [Jatropha curcas] |
| 9 |
Hb_000264_100 |
0.0764546572 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_025477_060 |
0.076947039 |
- |
- |
PREDICTED: calcium-dependent protein kinase 26-like [Jatropha curcas] |
| 11 |
Hb_000475_020 |
0.0770863193 |
- |
- |
PREDICTED: dehydrogenase/reductase SDR family member on chromosome X homolog isoform X2 [Jatropha curcas] |
| 12 |
Hb_000120_610 |
0.0775098996 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 13 |
Hb_056196_050 |
0.0778734125 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 4-like [Jatropha curcas] |
| 14 |
Hb_001080_270 |
0.078577775 |
- |
- |
PREDICTED: NF-kappa-B-activating protein [Jatropha curcas] |
| 15 |
Hb_033883_060 |
0.078958714 |
- |
- |
PREDICTED: uncharacterized protein LOC105648353 [Jatropha curcas] |
| 16 |
Hb_007904_180 |
0.0807461998 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 6-like [Jatropha curcas] |
| 17 |
Hb_000363_130 |
0.0810447849 |
- |
- |
PREDICTED: FRIGIDA-like protein 1 [Jatropha curcas] |
| 18 |
Hb_000640_200 |
0.0811295833 |
- |
- |
hypothetical protein RCOM_1346600 [Ricinus communis] |
| 19 |
Hb_000227_380 |
0.0812271801 |
- |
- |
PREDICTED: protein TIME FOR COFFEE isoform X3 [Jatropha curcas] |
| 20 |
Hb_004970_160 |
0.0812290931 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |