| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
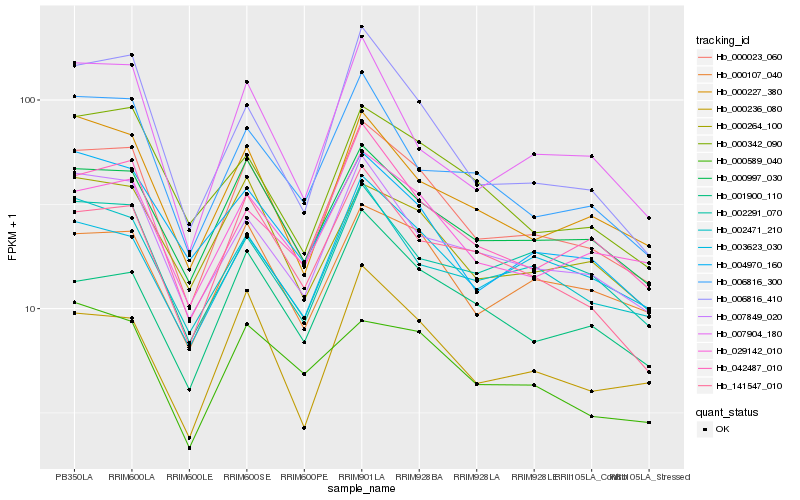
Hb_000023_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633775 [Jatropha curcas] |
| 2 |
Hb_000997_030 |
0.0686984806 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 3 |
Hb_029142_010 |
0.0717832648 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 4 |
Hb_007849_020 |
0.0743128915 |
transcription factor |
TF Family: MYB |
cell division control protein, putative [Ricinus communis] |
| 5 |
Hb_000107_040 |
0.0761467282 |
- |
- |
nuclear pore complex protein nup153, putative [Ricinus communis] |
| 6 |
Hb_000227_380 |
0.0776940196 |
- |
- |
PREDICTED: protein TIME FOR COFFEE isoform X3 [Jatropha curcas] |
| 7 |
Hb_042487_010 |
0.0810091102 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 9-like [Jatropha curcas] |
| 8 |
Hb_000236_080 |
0.0822828205 |
- |
- |
PREDICTED: small subunit processome component 20 homolog [Jatropha curcas] |
| 9 |
Hb_141547_010 |
0.0832370986 |
- |
- |
PREDICTED: ARF guanine-nucleotide exchange factor GNOM [Jatropha curcas] |
| 10 |
Hb_006816_300 |
0.0853960738 |
- |
- |
PREDICTED: importin subunit alpha-2 [Jatropha curcas] |
| 11 |
Hb_006816_410 |
0.0854572263 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit A [Jatropha curcas] |
| 12 |
Hb_002471_210 |
0.085662854 |
- |
- |
PREDICTED: THO complex subunit 2 [Jatropha curcas] |
| 13 |
Hb_000264_100 |
0.0858139018 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000342_090 |
0.0902784732 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001900_110 |
0.0915789843 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit E [Jatropha curcas] |
| 16 |
Hb_003623_030 |
0.0916695785 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 14 [Jatropha curcas] |
| 17 |
Hb_007904_180 |
0.0920043448 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 6-like [Jatropha curcas] |
| 18 |
Hb_004970_160 |
0.0928115092 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_002291_070 |
0.0935591965 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Prunus mume] |
| 20 |
Hb_000589_040 |
0.0947707554 |
desease resistance |
Gene Name: zf-BED |
PREDICTED: probable disease resistance protein At1g12280 [Jatropha curcas] |