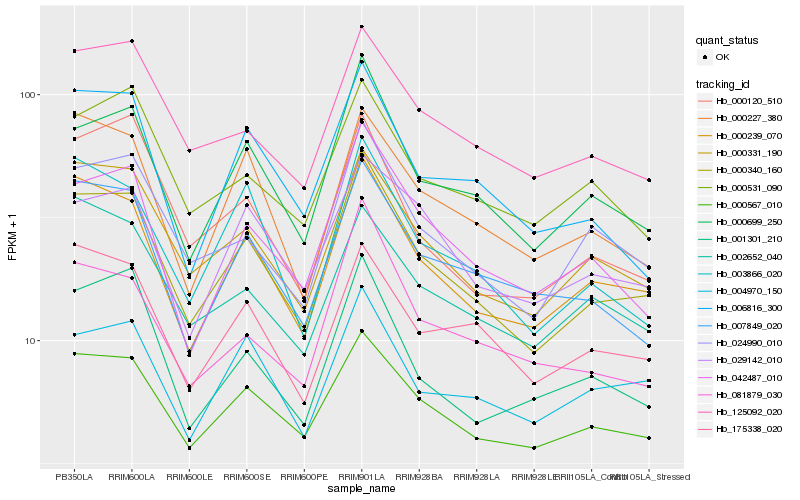
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_160 |
0.0 |
- |
- |
PREDICTED: elongation factor Tu, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000239_070 |
0.0524019141 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000699_250 |
0.0591817124 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 4 |
Hb_000331_190 |
0.0613429793 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 5 |
Hb_024990_010 |
0.070401864 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 6 |
Hb_000567_010 |
0.0738700125 |
- |
- |
PREDICTED: uncharacterized protein LOC105631110 [Jatropha curcas] |
| 7 |
Hb_125092_020 |
0.0777288919 |
- |
- |
PREDICTED: nucleolin 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_042487_010 |
0.0801038404 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 9-like [Jatropha curcas] |
| 9 |
Hb_003866_020 |
0.0812641 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_175338_020 |
0.0813368461 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 11 |
Hb_000227_380 |
0.0830318577 |
- |
- |
PREDICTED: protein TIME FOR COFFEE isoform X3 [Jatropha curcas] |
| 12 |
Hb_004970_150 |
0.0830840333 |
- |
- |
PREDICTED: uncharacterized protein LOC105633636 [Jatropha curcas] |
| 13 |
Hb_007849_020 |
0.0838211012 |
transcription factor |
TF Family: MYB |
cell division control protein, putative [Ricinus communis] |
| 14 |
Hb_001301_210 |
0.0848783845 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 32 [Jatropha curcas] |
| 15 |
Hb_029142_010 |
0.0864154242 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 16 |
Hb_081879_030 |
0.0879269308 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 21 homolog [Jatropha curcas] |
| 17 |
Hb_002652_040 |
0.0883623123 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 18 |
Hb_000120_510 |
0.089134197 |
- |
- |
PREDICTED: RNA polymerase I-specific transcription initiation factor RRN3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000531_090 |
0.0891685092 |
- |
- |
PREDICTED: transcription initiation factor IIF subunit alpha [Jatropha curcas] |
| 20 |
Hb_006816_300 |
0.0902285205 |
- |
- |
PREDICTED: importin subunit alpha-2 [Jatropha curcas] |