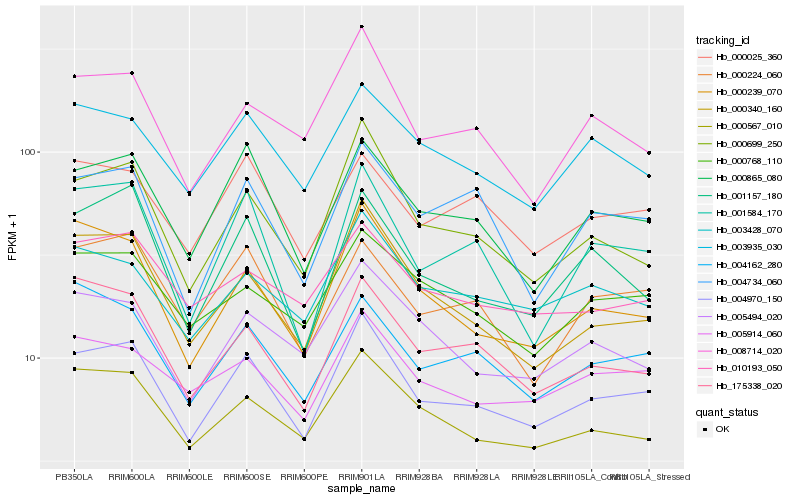
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004970_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633636 [Jatropha curcas] |
| 2 |
Hb_000865_080 |
0.0710149891 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 3 |
Hb_000699_250 |
0.0729811143 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 4 |
Hb_000567_010 |
0.0766017748 |
- |
- |
PREDICTED: uncharacterized protein LOC105631110 [Jatropha curcas] |
| 5 |
Hb_003428_070 |
0.0805667661 |
- |
- |
DEAD-box ATP-dependent RNA helicase 46 -like protein [Gossypium arboreum] |
| 6 |
Hb_003935_030 |
0.0806622037 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 7 |
Hb_005914_060 |
0.0820798954 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 8 |
Hb_000340_160 |
0.0830840333 |
- |
- |
PREDICTED: elongation factor Tu, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000025_360 |
0.0841061868 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000239_070 |
0.0842974583 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_175338_020 |
0.0857569877 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 12 |
Hb_008714_020 |
0.0858597126 |
- |
- |
PREDICTED: GTP-binding protein SAR1A-like [Jatropha curcas] |
| 13 |
Hb_000224_060 |
0.0861698808 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6 [Jatropha curcas] |
| 14 |
Hb_004162_280 |
0.0869173364 |
- |
- |
PREDICTED: uncharacterized protein LOC105628004 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000768_110 |
0.0871176541 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005494_020 |
0.0878245288 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 17 |
Hb_010193_050 |
0.0881659396 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001157_180 |
0.0893079883 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_004734_060 |
0.0894927213 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 32 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001584_170 |
0.0899979876 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |