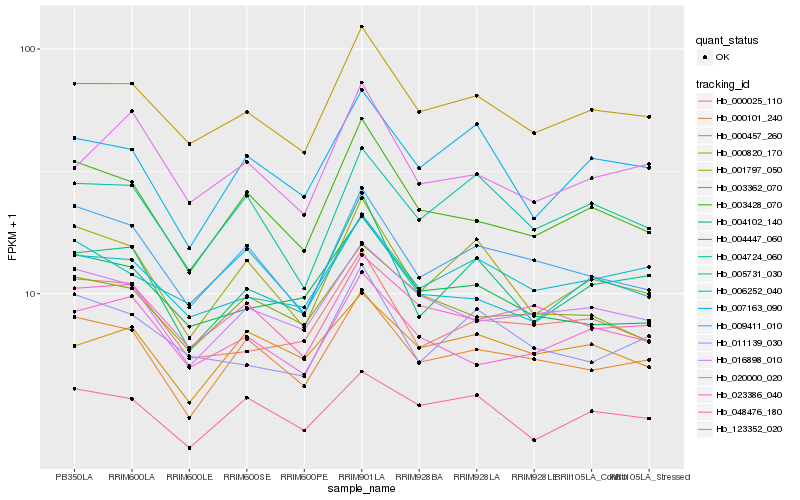
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000820_170 |
0.0 |
- |
- |
PREDICTED: THO complex subunit 4A [Jatropha curcas] |
| 2 |
Hb_006252_040 |
0.0473316812 |
- |
- |
queuine tRNA-ribosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_003428_070 |
0.0633145414 |
- |
- |
DEAD-box ATP-dependent RNA helicase 46 -like protein [Gossypium arboreum] |
| 4 |
Hb_016898_010 |
0.0647717733 |
- |
- |
PREDICTED: uncharacterized protein PFB0145c [Jatropha curcas] |
| 5 |
Hb_005731_030 |
0.0653495717 |
- |
- |
PREDICTED: UPF0400 protein C337.03 [Jatropha curcas] |
| 6 |
Hb_000101_240 |
0.0657722796 |
desease resistance |
Gene Name: NB-ARC |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 7 |
Hb_123352_020 |
0.0668937762 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g65560 [Jatropha curcas] |
| 8 |
Hb_020000_020 |
0.0670023167 |
- |
- |
Troponin C, skeletal muscle, putative [Ricinus communis] |
| 9 |
Hb_048476_180 |
0.0670494891 |
- |
- |
PREDICTED: nuclear pore complex protein NUP155 [Jatropha curcas] |
| 10 |
Hb_004724_060 |
0.0672186856 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004447_060 |
0.0676986594 |
- |
- |
PREDICTED: uncharacterized protein LOC105636329 isoform X2 [Jatropha curcas] |
| 12 |
Hb_003362_070 |
0.0679217798 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 13 |
Hb_004102_140 |
0.0680498972 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_023386_040 |
0.0685387688 |
- |
- |
PREDICTED: chaperone protein dnaJ 13-like [Populus euphratica] |
| 15 |
Hb_001797_050 |
0.0706200023 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_011139_030 |
0.0707234815 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR2-like [Jatropha curcas] |
| 17 |
Hb_007163_090 |
0.0708287684 |
- |
- |
PREDICTED: protein spt2-like [Jatropha curcas] |
| 18 |
Hb_000025_110 |
0.0708895191 |
- |
- |
hypothetical protein JCGZ_22722 [Jatropha curcas] |
| 19 |
Hb_000457_260 |
0.0724582059 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15690-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_009411_010 |
0.0725384469 |
- |
- |
PREDICTED: uncharacterized protein LOC105645272 [Jatropha curcas] |