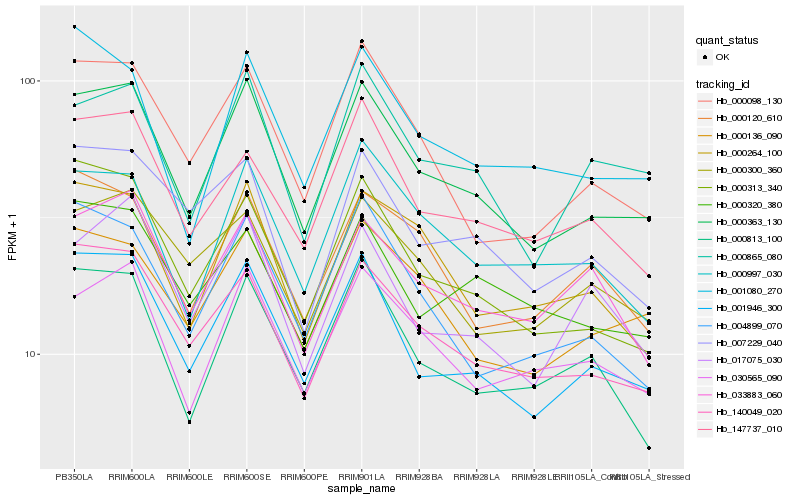
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000363_130 |
0.0 |
- |
- |
PREDICTED: FRIGIDA-like protein 1 [Jatropha curcas] |
| 2 |
Hb_001946_300 |
0.0502760863 |
- |
- |
PREDICTED: F-box protein At-B [Jatropha curcas] |
| 3 |
Hb_140049_020 |
0.0624895828 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 4 |
Hb_000313_340 |
0.0666820243 |
- |
- |
DNA repair protein xp-E, putative [Ricinus communis] |
| 5 |
Hb_000865_080 |
0.0693561071 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 6 |
Hb_030565_090 |
0.0703639618 |
- |
- |
PREDICTED: DNA repair protein RAD50 [Jatropha curcas] |
| 7 |
Hb_000136_090 |
0.0723495021 |
- |
- |
programmed cell death protein, putative [Ricinus communis] |
| 8 |
Hb_000098_130 |
0.0745344251 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2A [Jatropha curcas] |
| 9 |
Hb_147737_010 |
0.0769589192 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000997_030 |
0.0791298981 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 11 |
Hb_004899_070 |
0.0797964983 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 12 |
Hb_000264_100 |
0.0798061039 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000813_100 |
0.0810447849 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001080_270 |
0.0831686994 |
- |
- |
PREDICTED: NF-kappa-B-activating protein [Jatropha curcas] |
| 15 |
Hb_000300_360 |
0.0849951189 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 16 |
Hb_017075_030 |
0.0850140144 |
- |
- |
helicase, putative [Ricinus communis] |
| 17 |
Hb_033883_060 |
0.0855429678 |
- |
- |
PREDICTED: uncharacterized protein LOC105648353 [Jatropha curcas] |
| 18 |
Hb_007229_040 |
0.0859594377 |
- |
- |
PREDICTED: protein ARABIDILLO 1-like [Jatropha curcas] |
| 19 |
Hb_000120_610 |
0.0871285063 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 20 |
Hb_000320_380 |
0.0875197098 |
- |
- |
PREDICTED: uncharacterized protein LOC105649330 isoform X2 [Jatropha curcas] |