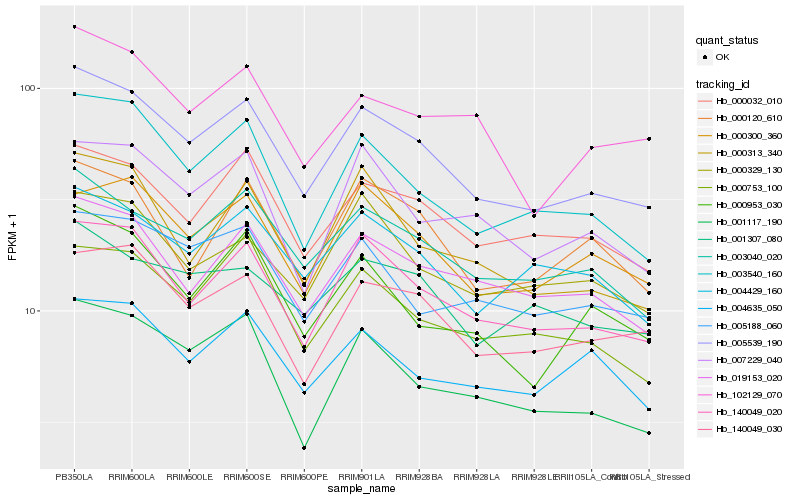
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005539_190 |
0.0 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 2 |
Hb_140049_020 |
0.0542513949 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 3 |
Hb_003040_020 |
0.0609525652 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 4 |
Hb_003540_160 |
0.0629716929 |
- |
- |
hypothetical protein POPTR_0011s12160g [Populus trichocarpa] |
| 5 |
Hb_000032_010 |
0.0673188713 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 6 |
Hb_019153_020 |
0.0707121353 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 7 |
Hb_140049_030 |
0.0741554263 |
- |
- |
non-specific lipid-transfer protein-like protein At2g13820 precursor [Jatropha curcas] |
| 8 |
Hb_004429_160 |
0.0748284897 |
- |
- |
PREDICTED: serine/threonine-protein kinase TAO3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001117_190 |
0.0776788693 |
transcription factor |
TF Family: FAR1 |
FAR1-related sequence 11 [Theobroma cacao] |
| 10 |
Hb_000753_100 |
0.0778058427 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640677 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000313_340 |
0.0778186386 |
- |
- |
DNA repair protein xp-E, putative [Ricinus communis] |
| 12 |
Hb_000300_360 |
0.079174596 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 13 |
Hb_005188_060 |
0.0808105284 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 14 |
Hb_000953_030 |
0.0811215787 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_001307_080 |
0.0822886813 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 16 |
Hb_102129_070 |
0.0830956406 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 124 [Cucumis melo] |
| 17 |
Hb_000329_130 |
0.0833404836 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 18 |
Hb_007229_040 |
0.0840594332 |
- |
- |
PREDICTED: protein ARABIDILLO 1-like [Jatropha curcas] |
| 19 |
Hb_004635_050 |
0.0845487833 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 20 |
Hb_000120_610 |
0.0847655772 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |