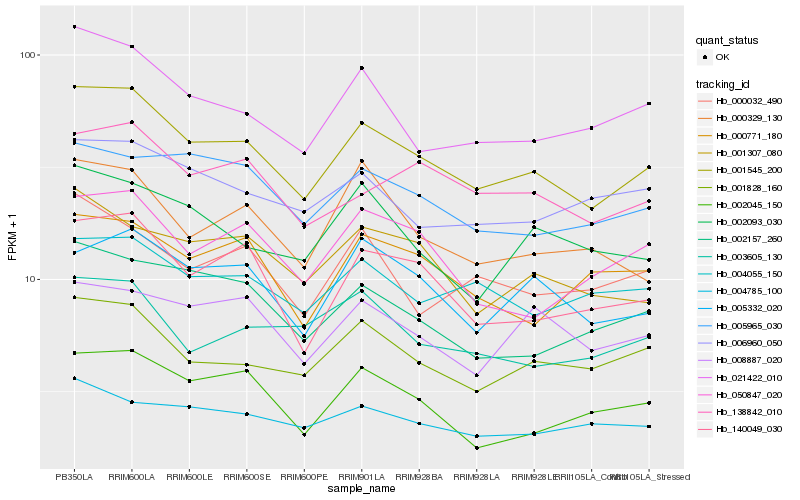
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001545_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized oxidoreductase At4g09670 [Jatropha curcas] |
| 2 |
Hb_001828_160 |
0.0724464399 |
- |
- |
hypothetical protein F775_31105 [Aegilops tauschii] |
| 3 |
Hb_140049_030 |
0.0741410951 |
- |
- |
non-specific lipid-transfer protein-like protein At2g13820 precursor [Jatropha curcas] |
| 4 |
Hb_021422_010 |
0.075723412 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 1, mitochondrial [Jatropha curcas] |
| 5 |
Hb_002157_260 |
0.0801473456 |
- |
- |
PREDICTED: partner of Y14 and mago-like [Jatropha curcas] |
| 6 |
Hb_005332_020 |
0.081141305 |
transcription factor |
TF Family: SNF2 |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 7 |
Hb_001307_080 |
0.0813588921 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 8 |
Hb_138842_010 |
0.0823578525 |
transcription factor |
TF Family: Orphans |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 9 |
Hb_005965_030 |
0.083531068 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 10 |
Hb_004055_150 |
0.086059008 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002045_150 |
0.0874986129 |
- |
- |
histone acetyltransferase gcn5, putative [Ricinus communis] |
| 12 |
Hb_050847_020 |
0.0879532611 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 13 |
Hb_006960_050 |
0.0889197858 |
- |
- |
PREDICTED: PRKR-interacting protein 1 [Jatropha curcas] |
| 14 |
Hb_008887_020 |
0.0895797325 |
- |
- |
PREDICTED: uncharacterized protein LOC105636191 [Jatropha curcas] |
| 15 |
Hb_002093_030 |
0.0901069558 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 16 |
Hb_000032_490 |
0.0911209877 |
- |
- |
PREDICTED: uncharacterized protein At4g10930-like [Jatropha curcas] |
| 17 |
Hb_004785_100 |
0.0914885173 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000329_130 |
0.0916880594 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 19 |
Hb_000771_180 |
0.0917709188 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 20 |
Hb_003605_130 |
0.0918771851 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Populus euphratica] |