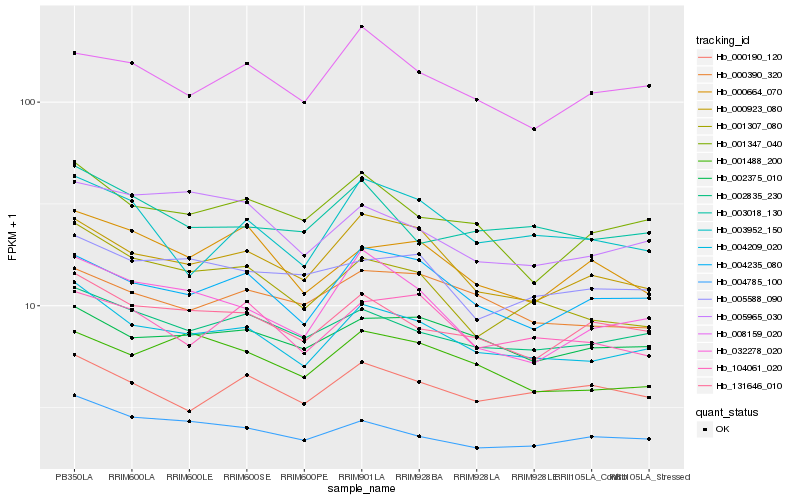
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004209_020 |
0.0 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 51-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_002835_230 |
0.0616078149 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_131646_010 |
0.0638481208 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_004235_080 |
0.0643101486 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 5 |
Hb_000923_080 |
0.0644012773 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2A [Jatropha curcas] |
| 6 |
Hb_004785_100 |
0.0679497671 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002375_010 |
0.0716599239 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001307_080 |
0.0724657349 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 9 |
Hb_001347_040 |
0.0746678411 |
- |
- |
PREDICTED: uncharacterized protein LOC105633920 [Jatropha curcas] |
| 10 |
Hb_104061_020 |
0.0762582037 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |
| 11 |
Hb_000390_320 |
0.0783744798 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-1b-like [Jatropha curcas] |
| 12 |
Hb_003018_130 |
0.0793771894 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 13 |
Hb_001488_200 |
0.079381447 |
- |
- |
PREDICTED: ribosome biogenesis protein WDR12 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_000190_120 |
0.0804342261 |
- |
- |
PREDICTED: uncharacterized protein LOC105649936 [Jatropha curcas] |
| 15 |
Hb_032278_020 |
0.081332332 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 16 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005588_090 |
0.0818291473 |
- |
- |
hypothetical protein CICLE_v10003480mg [Citrus clementina] |
| 17 |
Hb_008159_020 |
0.0837599156 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 18 |
Hb_003952_150 |
0.0843095218 |
- |
- |
PREDICTED: cactin [Jatropha curcas] |
| 19 |
Hb_005965_030 |
0.0849304713 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 20 |
Hb_000664_070 |
0.0849903921 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |