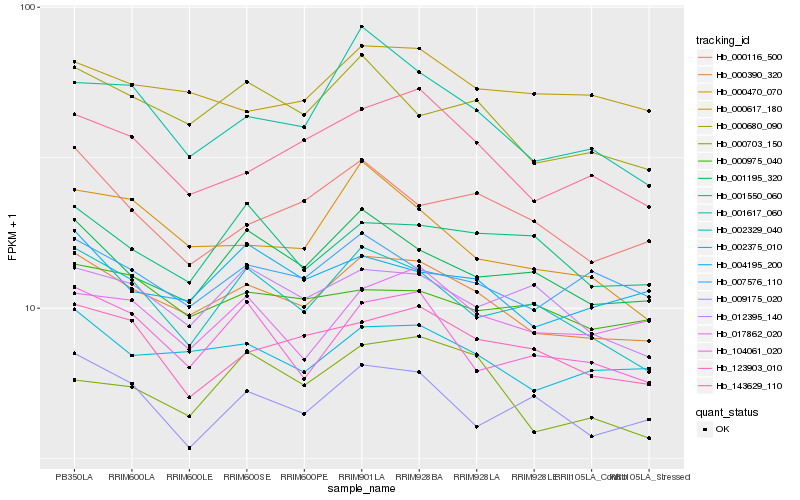
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000390_320 |
0.0 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-1b-like [Jatropha curcas] |
| 2 |
Hb_002375_010 |
0.0472735646 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000680_090 |
0.0539788917 |
- |
- |
Protein SEY1, putative [Ricinus communis] |
| 4 |
Hb_001195_320 |
0.0596085599 |
- |
- |
PREDICTED: S-adenosyl-L-methionine-dependent tRNA 4-demethylwyosine synthase [Jatropha curcas] |
| 5 |
Hb_143629_110 |
0.0596192689 |
- |
- |
glutamyl-tRNA synthetase, cytoplasmic, putative [Ricinus communis] |
| 6 |
Hb_002329_040 |
0.0628419416 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 7 |
Hb_123903_010 |
0.0632458211 |
transcription factor |
TF Family: IWS1 |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004195_200 |
0.0638047326 |
- |
- |
glycosyl transferase family 1 family protein [Populus trichocarpa] |
| 9 |
Hb_007576_110 |
0.0650479419 |
- |
- |
Endoplasmic reticulum vesicle transporter protein [Theobroma cacao] |
| 10 |
Hb_012395_140 |
0.0662439054 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 11 |
Hb_000617_180 |
0.0681574016 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 12 |
Hb_000975_040 |
0.0689265783 |
- |
- |
hypothetical protein CICLE_v10028249mg [Citrus clementina] |
| 13 |
Hb_104061_020 |
0.0690482156 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |
| 14 |
Hb_001617_060 |
0.0697876664 |
- |
- |
PREDICTED: polyadenylate-binding protein RBP45B isoform X1 [Jatropha curcas] |
| 15 |
Hb_001550_060 |
0.0699639443 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X4 [Jatropha curcas] |
| 16 |
Hb_017862_020 |
0.0706344477 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase lvsG [Jatropha curcas] |
| 17 |
Hb_009175_020 |
0.0708636125 |
- |
- |
PREDICTED: uncharacterized protein LOC101303140 [Fragaria vesca subsp. vesca] |
| 18 |
Hb_000116_500 |
0.0715211609 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 11 [Jatropha curcas] |
| 19 |
Hb_000703_150 |
0.0717612745 |
- |
- |
PREDICTED: F-box/LRR-repeat MAX2 homolog A [Jatropha curcas] |
| 20 |
Hb_000470_070 |
0.0718263814 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |