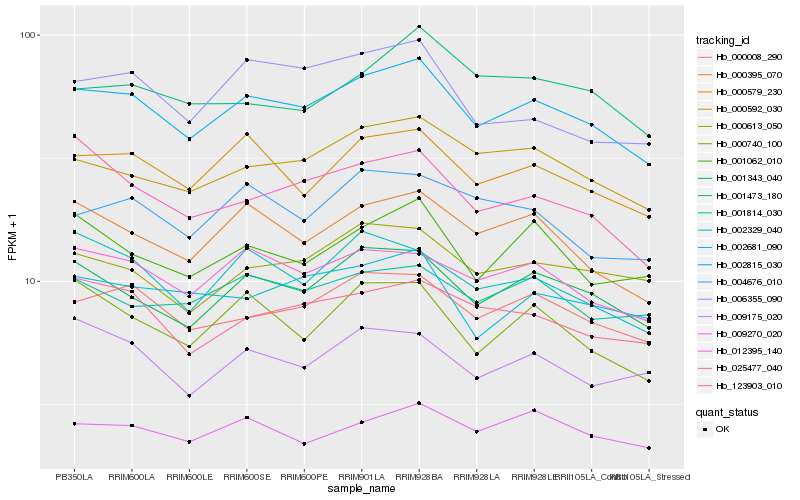
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002815_030 |
0.0 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 2 |
Hb_001343_040 |
0.0459409893 |
- |
- |
PREDICTED: uncharacterized protein LOC105638555 [Jatropha curcas] |
| 3 |
Hb_000008_290 |
0.047090523 |
- |
- |
PREDICTED: protein SAND [Jatropha curcas] |
| 4 |
Hb_000579_230 |
0.0492785138 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 5 |
Hb_012395_140 |
0.0518353541 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 6 |
Hb_000395_070 |
0.0543402931 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000592_030 |
0.055278616 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 8 |
Hb_009270_020 |
0.062042561 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: helicase and polymerase-containing protein TEBICHI [Jatropha curcas] |
| 9 |
Hb_123903_010 |
0.0620542616 |
transcription factor |
TF Family: IWS1 |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000613_050 |
0.0623938482 |
- |
- |
PREDICTED: uncharacterized protein LOC105641544 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001062_010 |
0.0639435064 |
- |
- |
PREDICTED: CWF19-like protein 2 [Jatropha curcas] |
| 12 |
Hb_001814_030 |
0.0643352604 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 13 |
Hb_002681_090 |
0.06514699 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002329_040 |
0.0652159619 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 15 |
Hb_000740_100 |
0.0663725594 |
- |
- |
calpain, putative [Ricinus communis] |
| 16 |
Hb_004676_010 |
0.067783024 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g80270, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_025477_040 |
0.0680545136 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 [Jatropha curcas] |
| 18 |
Hb_006355_090 |
0.0696060655 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S proteasome regulatory subunit 4 homolog A [Jatropha curcas] |
| 19 |
Hb_009175_020 |
0.0703942278 |
- |
- |
PREDICTED: uncharacterized protein LOC101303140 [Fragaria vesca subsp. vesca] |
| 20 |
Hb_001473_180 |
0.0727981796 |
- |
- |
glutathione reductase [Hevea brasiliensis] |