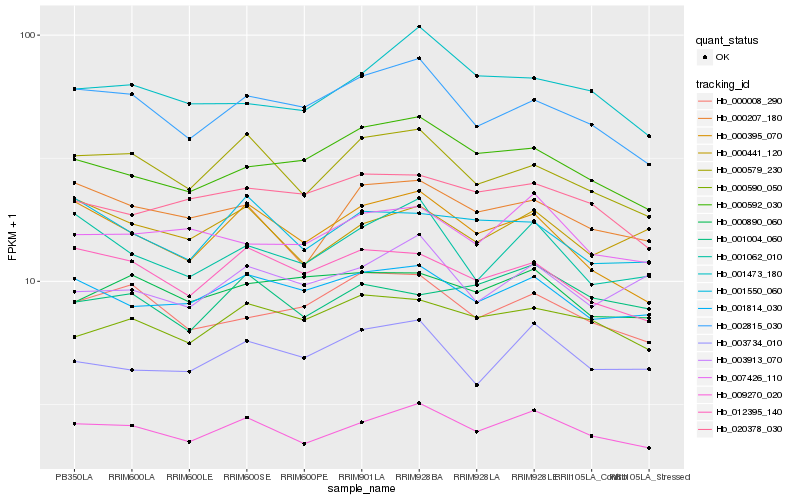
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009270_020 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: helicase and polymerase-containing protein TEBICHI [Jatropha curcas] |
| 2 |
Hb_001814_030 |
0.0541996782 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 3 |
Hb_000579_230 |
0.0560252845 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 4 |
Hb_000207_180 |
0.0585052987 |
- |
- |
PREDICTED: calponin homology domain-containing protein DDB_G0272472 [Jatropha curcas] |
| 5 |
Hb_007426_110 |
0.0601630988 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 6 |
Hb_000590_050 |
0.0606128452 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 7 |
Hb_000441_120 |
0.0609108259 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT111, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_002815_030 |
0.062042561 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 9 |
Hb_012395_140 |
0.0635074641 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 10 |
Hb_003734_010 |
0.0654781084 |
- |
- |
PREDICTED: double-strand break repair protein MRE11 [Jatropha curcas] |
| 11 |
Hb_001473_180 |
0.0656614491 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 12 |
Hb_001004_060 |
0.0657749015 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 13 |
Hb_020378_030 |
0.0663202872 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 14 |
Hb_000592_030 |
0.0671355132 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 15 |
Hb_001550_060 |
0.0681443404 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X4 [Jatropha curcas] |
| 16 |
Hb_001062_010 |
0.0684582711 |
- |
- |
PREDICTED: CWF19-like protein 2 [Jatropha curcas] |
| 17 |
Hb_000890_060 |
0.0684833585 |
- |
- |
PREDICTED: sucrose nonfermenting 4-like protein [Jatropha curcas] |
| 18 |
Hb_000395_070 |
0.0691092713 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003913_070 |
0.0692668706 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 30 [Populus euphratica] |
| 20 |
Hb_000008_290 |
0.0693164701 |
- |
- |
PREDICTED: protein SAND [Jatropha curcas] |