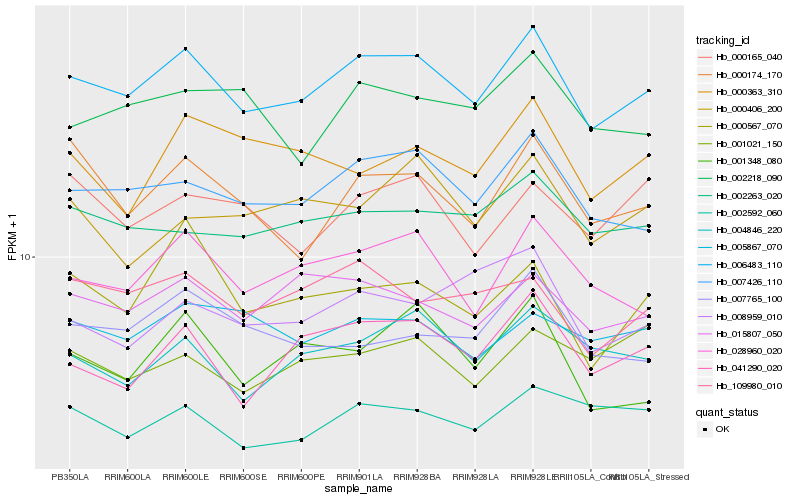
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006483_110 |
0.0 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 2 |
Hb_007426_110 |
0.0447074082 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 3 |
Hb_028960_020 |
0.0557182256 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 4 |
Hb_015807_050 |
0.0573255095 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 5 |
Hb_041290_020 |
0.0582407143 |
- |
- |
PREDICTED: centromere/kinetochore protein zw10 homolog isoform X2 [Jatropha curcas] |
| 6 |
Hb_002263_020 |
0.0614548565 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 7 |
Hb_109980_010 |
0.0618525502 |
- |
- |
PREDICTED: uncharacterized protein LOC105647182 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004846_220 |
0.0625223822 |
- |
- |
PREDICTED: probable protein phosphatase 2C 11 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002592_060 |
0.0638415109 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000174_170 |
0.0652582254 |
- |
- |
PREDICTED: splicing factor U2AF-associated protein 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005867_070 |
0.0661992942 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_007765_100 |
0.0671426737 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 13 |
Hb_002218_090 |
0.0673398167 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 14 |
Hb_000567_070 |
0.0676030229 |
- |
- |
autophagy protein, putative [Ricinus communis] |
| 15 |
Hb_001348_080 |
0.0680905168 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 16 |
Hb_000165_040 |
0.0714787489 |
- |
- |
hypothetical protein JCGZ_01206 [Jatropha curcas] |
| 17 |
Hb_001021_150 |
0.0717953294 |
- |
- |
PREDICTED: metal tolerance protein C1 [Jatropha curcas] |
| 18 |
Hb_000363_310 |
0.0722419557 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 19 |
Hb_008959_010 |
0.072345372 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000406_200 |
0.0724859368 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit epsilon-like isoform X4 [Jatropha curcas] |