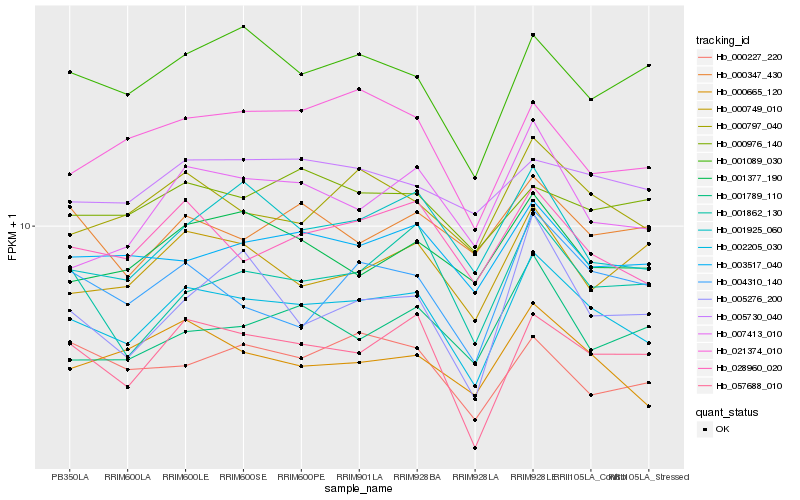
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002205_030 |
0.0 |
- |
- |
PREDICTED: alpha-galactosidase 3 [Jatropha curcas] |
| 2 |
Hb_057688_010 |
0.0565528196 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X2 [Jatropha curcas] |
| 3 |
Hb_028960_020 |
0.0581327755 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000797_040 |
0.0608043644 |
- |
- |
PREDICTED: uncharacterized protein LOC105645914 [Jatropha curcas] |
| 5 |
Hb_003517_040 |
0.0652067534 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000665_120 |
0.0656827601 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105637593 [Jatropha curcas] |
| 7 |
Hb_001377_190 |
0.070838436 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 8 |
Hb_007413_010 |
0.0710786828 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 9 |
Hb_001089_030 |
0.0712608884 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 10 |
Hb_001925_060 |
0.0720846345 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |
| 11 |
Hb_005730_040 |
0.0745965753 |
- |
- |
hypothetical protein PRUPE_ppa004338mg [Prunus persica] |
| 12 |
Hb_004310_140 |
0.0749304655 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_021374_010 |
0.0749345049 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-A1 [Populus euphratica] |
| 14 |
Hb_000749_010 |
0.0749786127 |
- |
- |
PREDICTED: non-canonical poly(A) RNA polymerase PAPD5 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001789_110 |
0.0761767985 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 16 |
Hb_001862_130 |
0.0770430589 |
- |
- |
Protein FRIGIDA, putative [Ricinus communis] |
| 17 |
Hb_000976_140 |
0.0779316401 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000227_220 |
0.0781277154 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 19 |
Hb_000347_430 |
0.0784076503 |
- |
- |
PREDICTED: mitochondrial ubiquitin ligase activator of nfkb 1-A [Jatropha curcas] |
| 20 |
Hb_005276_200 |
0.0791399366 |
- |
- |
PREDICTED: preprotein translocase subunit SCY2, chloroplastic [Jatropha curcas] |