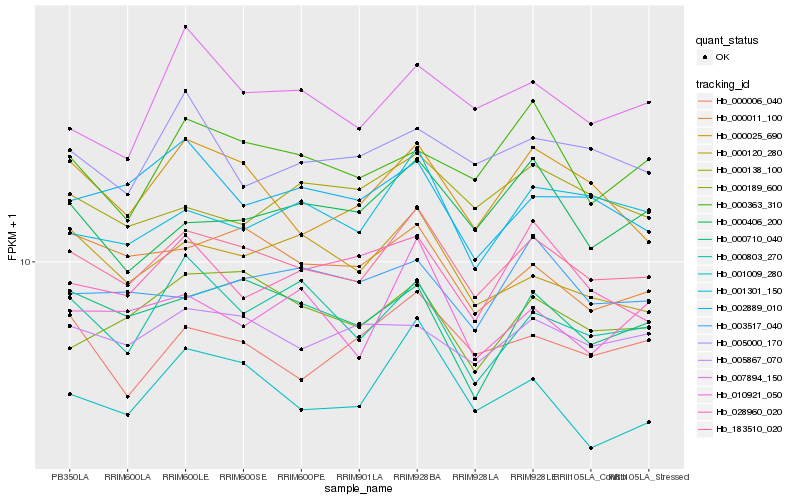
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_183510_020 |
0.0 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 2 |
Hb_000025_690 |
0.0572487597 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 3 |
Hb_005867_070 |
0.0577318751 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_000011_100 |
0.0616784267 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 5 |
Hb_000006_040 |
0.0619825211 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 6 |
Hb_000710_040 |
0.0632777421 |
- |
- |
PREDICTED: HEAT repeat-containing protein 6 [Jatropha curcas] |
| 7 |
Hb_001301_150 |
0.0636891615 |
- |
- |
PREDICTED: UPF0136 membrane protein At2g26240 [Vitis vinifera] |
| 8 |
Hb_000120_280 |
0.0642037458 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 9 |
Hb_000189_600 |
0.0642234543 |
- |
- |
PREDICTED: protein MON2 homolog isoform X1 [Jatropha curcas] |
| 10 |
Hb_001009_280 |
0.0646614755 |
- |
- |
PREDICTED: telomere length regulation protein TEL2 homolog [Jatropha curcas] |
| 11 |
Hb_002889_010 |
0.0676688342 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 12 |
Hb_000406_200 |
0.0688017122 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit epsilon-like isoform X4 [Jatropha curcas] |
| 13 |
Hb_000803_270 |
0.0690183447 |
- |
- |
PREDICTED: nuclear cap-binding protein subunit 1 [Jatropha curcas] |
| 14 |
Hb_000138_100 |
0.0699014016 |
- |
- |
bifunctional purine biosynthesis protein, putative [Ricinus communis] |
| 15 |
Hb_007894_150 |
0.0699077608 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 16 |
Hb_003517_040 |
0.0699114511 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000363_310 |
0.0700417141 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 18 |
Hb_010921_050 |
0.0702434151 |
- |
- |
PREDICTED: protein POLLEN DEFECTIVE IN GUIDANCE 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_028960_020 |
0.0702899854 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005000_170 |
0.0704105302 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |