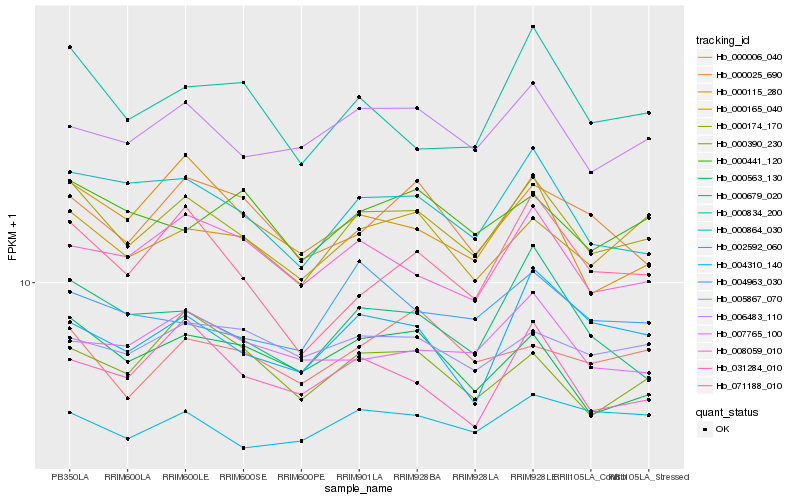
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000174_170 |
0.0 |
- |
- |
PREDICTED: splicing factor U2AF-associated protein 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000864_030 |
0.0590085886 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1XR1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000165_040 |
0.0640096316 |
- |
- |
hypothetical protein JCGZ_01206 [Jatropha curcas] |
| 4 |
Hb_006483_110 |
0.0652582254 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 5 |
Hb_000679_020 |
0.072429687 |
transcription factor |
TF Family: HB |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 6 |
Hb_000563_130 |
0.0729558231 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 7 |
Hb_008059_010 |
0.0729940782 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 8 |
Hb_071188_010 |
0.073546663 |
- |
- |
hypothetical protein POPTR_0013s025001g, partial [Populus trichocarpa] |
| 9 |
Hb_002592_060 |
0.0768204764 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000834_200 |
0.0772882864 |
- |
- |
PREDICTED: splicing factor, arginine/serine-rich 19 [Jatropha curcas] |
| 11 |
Hb_000025_690 |
0.0776768368 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 12 |
Hb_004310_140 |
0.0779064512 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_000390_230 |
0.0786915666 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 14 |
Hb_000441_120 |
0.0787438676 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT111, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_031284_010 |
0.0790998847 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 16 |
Hb_000115_280 |
0.0791110944 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 17 |
Hb_005867_070 |
0.0797522414 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_007765_100 |
0.0809474454 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 19 |
Hb_000006_040 |
0.0815023268 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 20 |
Hb_004963_030 |
0.0816124256 |
- |
- |
PREDICTED: twinkle homolog protein, chloroplastic/mitochondrial [Jatropha curcas] |