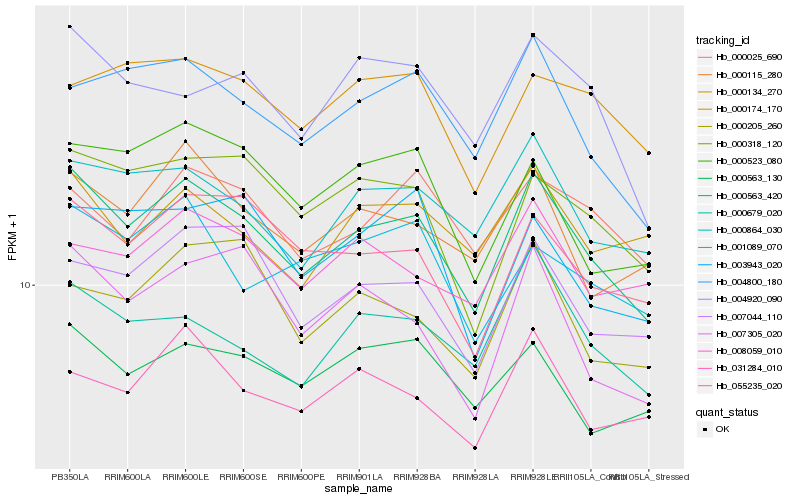
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000563_420 |
0.0 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 2 |
Hb_000864_030 |
0.0674773306 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1XR1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_004800_180 |
0.0702165211 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 4 |
Hb_055235_020 |
0.0717019586 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000115_280 |
0.0724068582 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 6 |
Hb_000318_120 |
0.0728356378 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: DNA ligase 1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_007044_110 |
0.0737196491 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_000679_020 |
0.0762219756 |
transcription factor |
TF Family: HB |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 9 |
Hb_004920_090 |
0.0769731079 |
- |
- |
PREDICTED: probable glutamyl endopeptidase, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_000563_130 |
0.0785036744 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 11 |
Hb_000523_080 |
0.0794863794 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 12 |
Hb_031284_010 |
0.0797265222 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 13 |
Hb_007305_020 |
0.0799288881 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 14 |
Hb_000025_690 |
0.0812222071 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 15 |
Hb_001089_070 |
0.0824106323 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 16 |
Hb_000174_170 |
0.0840399253 |
- |
- |
PREDICTED: splicing factor U2AF-associated protein 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000205_260 |
0.0840940035 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 18 |
Hb_008059_010 |
0.0849537146 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000134_270 |
0.0857498855 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 20 |
Hb_003943_020 |
0.08596472 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |