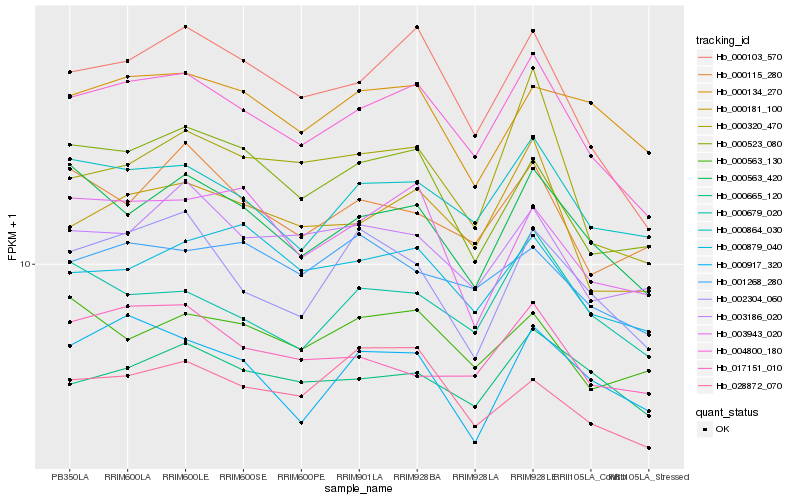
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004800_180 |
0.0 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 2 |
Hb_000103_570 |
0.0575437015 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000181_100 |
0.0606929265 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |
| 4 |
Hb_000864_030 |
0.0636542428 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1XR1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000563_420 |
0.0702165211 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 6 |
Hb_000665_120 |
0.0799921583 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105637593 [Jatropha curcas] |
| 7 |
Hb_000679_020 |
0.0819346598 |
transcription factor |
TF Family: HB |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 8 |
Hb_000523_080 |
0.0823348686 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 9 |
Hb_003186_020 |
0.0834934394 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000879_040 |
0.0836297702 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 11 |
Hb_017151_010 |
0.0845842875 |
- |
- |
PREDICTED: uncharacterized protein LOC105641966 [Jatropha curcas] |
| 12 |
Hb_002304_060 |
0.0856431224 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 13 |
Hb_000917_320 |
0.0867994036 |
- |
- |
PREDICTED: uncharacterized protein LOC105641975 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001268_280 |
0.087202074 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000320_470 |
0.0875211207 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 16 |
Hb_000115_280 |
0.0877376366 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 17 |
Hb_000134_270 |
0.0879433636 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 18 |
Hb_028872_070 |
0.0888936309 |
desease resistance |
Gene Name: DEAD |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase PRP1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000563_130 |
0.0894678788 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 20 |
Hb_003943_020 |
0.0894828747 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |