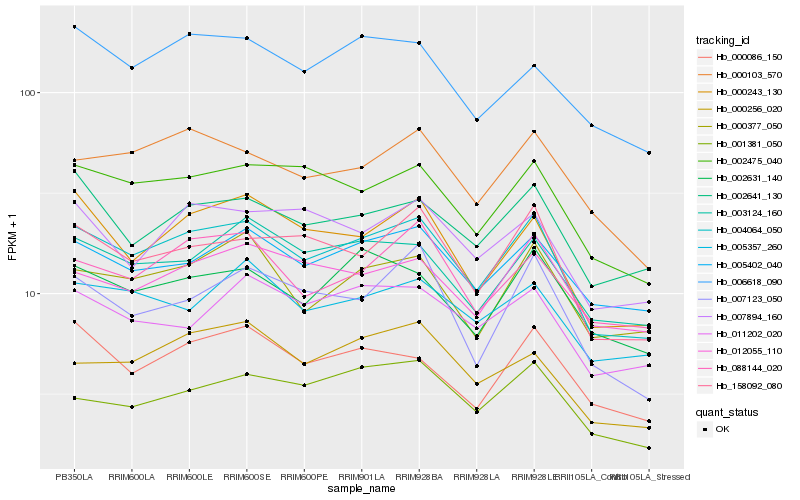
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004064_050 |
0.0 |
- |
- |
Structural maintenance of chromosome, putative [Ricinus communis] |
| 2 |
Hb_002475_040 |
0.0675391381 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 3 |
Hb_158092_080 |
0.0681707655 |
- |
- |
hypothetical protein JCGZ_23092 [Jatropha curcas] |
| 4 |
Hb_000243_130 |
0.07295725 |
- |
- |
hypothetical protein M569_10795, partial [Genlisea aurea] |
| 5 |
Hb_005402_040 |
0.0741667516 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 6 |
Hb_003124_160 |
0.0751748009 |
- |
- |
dynamin, putative [Ricinus communis] |
| 7 |
Hb_000377_050 |
0.0777864553 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 8 |
Hb_000103_570 |
0.0779267598 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002631_140 |
0.0791862446 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 12, chloroplastic [Jatropha curcas] |
| 10 |
Hb_012055_110 |
0.0797793123 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 11 |
Hb_002641_130 |
0.0803982958 |
- |
- |
PREDICTED: uncharacterized protein LOC105643556 [Jatropha curcas] |
| 12 |
Hb_000086_150 |
0.0805402346 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |
| 13 |
Hb_088144_020 |
0.0813256867 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000256_020 |
0.0829269401 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 15 |
Hb_005357_260 |
0.082998347 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 16 |
Hb_001381_050 |
0.0838448994 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 17 |
Hb_007123_050 |
0.085116391 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 18 |
Hb_006618_090 |
0.0852594965 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 19 |
Hb_007894_160 |
0.085729465 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 20 |
Hb_011202_020 |
0.0863587304 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |