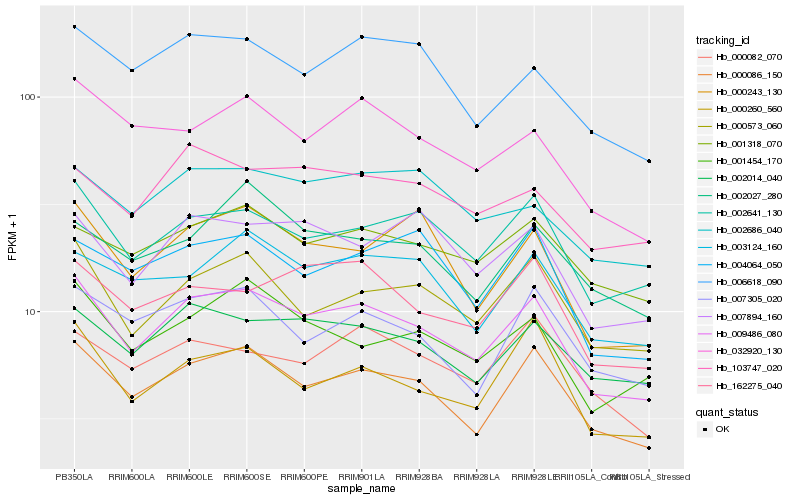
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009486_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643242 [Jatropha curcas] |
| 2 |
Hb_000086_150 |
0.0516076972 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002641_130 |
0.0702450183 |
- |
- |
PREDICTED: uncharacterized protein LOC105643556 [Jatropha curcas] |
| 4 |
Hb_000573_060 |
0.074221874 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2D isoform X1 [Jatropha curcas] |
| 5 |
Hb_002014_040 |
0.0762668605 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 6 |
Hb_001454_170 |
0.082302081 |
- |
- |
PREDICTED: uncharacterized protein LOC105643431 [Jatropha curcas] |
| 7 |
Hb_000260_560 |
0.0828569409 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18110, chloroplastic [Jatropha curcas] |
| 8 |
Hb_006618_090 |
0.0833010409 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 9 |
Hb_032920_130 |
0.0844183509 |
- |
- |
PREDICTED: vacuolar-sorting receptor 3 [Jatropha curcas] |
| 10 |
Hb_162275_040 |
0.084705403 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003124_160 |
0.0851145458 |
- |
- |
dynamin, putative [Ricinus communis] |
| 12 |
Hb_007305_020 |
0.0854420073 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 13 |
Hb_000243_130 |
0.0856189017 |
- |
- |
hypothetical protein M569_10795, partial [Genlisea aurea] |
| 14 |
Hb_001318_070 |
0.087990754 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007894_160 |
0.0882424037 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 16 |
Hb_002686_040 |
0.0915041319 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 17 |
Hb_004064_050 |
0.0915396447 |
- |
- |
Structural maintenance of chromosome, putative [Ricinus communis] |
| 18 |
Hb_000082_070 |
0.0917734041 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 1 [Jatropha curcas] |
| 19 |
Hb_103747_020 |
0.0938446855 |
- |
- |
calcium dependent protein kinase, partial [Hevea brasiliensis] |
| 20 |
Hb_002027_280 |
0.0941343016 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |