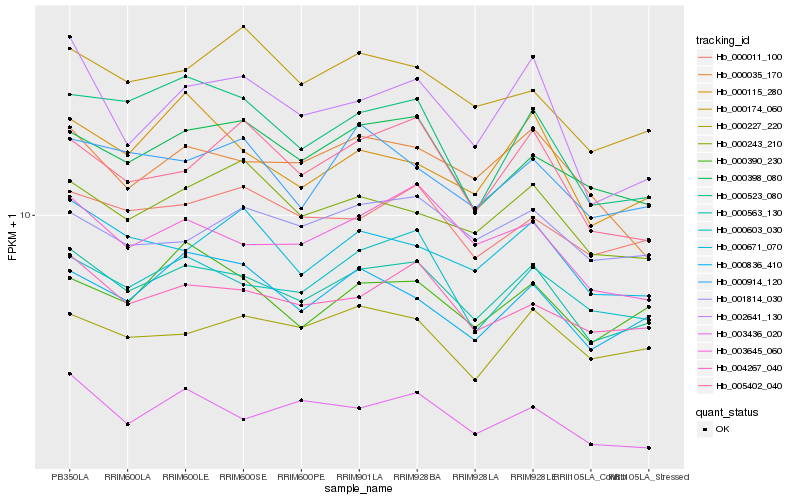
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000563_130 |
0.0 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 2 |
Hb_004267_040 |
0.0518571062 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |
| 3 |
Hb_003645_060 |
0.0574514877 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000603_030 |
0.0579757324 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 5 |
Hb_000398_080 |
0.0591844314 |
- |
- |
tip120, putative [Ricinus communis] |
| 6 |
Hb_000011_100 |
0.0607722218 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 7 |
Hb_000836_410 |
0.0632746153 |
- |
- |
sec10, putative [Ricinus communis] |
| 8 |
Hb_000671_070 |
0.063732272 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 9 |
Hb_000227_220 |
0.0641379853 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 10 |
Hb_000243_210 |
0.0659642899 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_000390_230 |
0.0664522931 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 12 |
Hb_000914_120 |
0.0669526126 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003436_020 |
0.0692138331 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000035_170 |
0.0694643581 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 2-like [Jatropha curcas] |
| 15 |
Hb_005402_040 |
0.0697461837 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 16 |
Hb_002641_130 |
0.0699897892 |
- |
- |
PREDICTED: uncharacterized protein LOC105643556 [Jatropha curcas] |
| 17 |
Hb_000523_080 |
0.07006213 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 18 |
Hb_001814_030 |
0.0703095494 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 19 |
Hb_000174_060 |
0.0707863975 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 20 |
Hb_000115_280 |
0.0716461521 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |