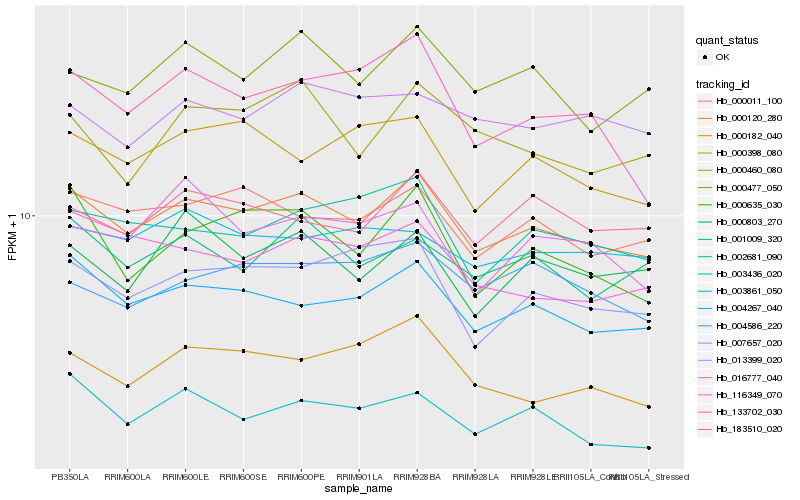
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_280 |
0.0 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 2 |
Hb_004267_040 |
0.0486933041 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |
| 3 |
Hb_000011_100 |
0.0561589918 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 4 |
Hb_000477_050 |
0.060442947 |
- |
- |
PREDICTED: 26S protease regulatory subunit 8 homolog A [Jatropha curcas] |
| 5 |
Hb_183510_020 |
0.0642037458 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 6 |
Hb_001009_320 |
0.0646281618 |
- |
- |
PREDICTED: uncharacterized protein LOC105640250 [Jatropha curcas] |
| 7 |
Hb_002681_090 |
0.0646889924 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 8 |
Hb_007657_020 |
0.065078589 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 9 |
Hb_000635_030 |
0.0650826066 |
- |
- |
mannosyl-oligosaccharide glucosidase, putative [Ricinus communis] |
| 10 |
Hb_000398_080 |
0.0680866473 |
- |
- |
tip120, putative [Ricinus communis] |
| 11 |
Hb_000803_270 |
0.0691768396 |
- |
- |
PREDICTED: nuclear cap-binding protein subunit 1 [Jatropha curcas] |
| 12 |
Hb_000460_080 |
0.0692104558 |
- |
- |
acyl-coenzyme A binding domain containing, putative [Ricinus communis] |
| 13 |
Hb_116349_070 |
0.0696865173 |
- |
- |
hypothetical protein JCGZ_21753 [Jatropha curcas] |
| 14 |
Hb_133702_030 |
0.0702777954 |
- |
- |
PREDICTED: uric acid degradation bifunctional protein TTL isoform X1 [Jatropha curcas] |
| 15 |
Hb_003861_050 |
0.0708075166 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 16 |
Hb_004586_220 |
0.07089353 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 17 |
Hb_013399_020 |
0.0709843666 |
- |
- |
PREDICTED: developmentally-regulated G-protein 2 [Jatropha curcas] |
| 18 |
Hb_000182_040 |
0.0730122095 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein CHROMATIN REMODELING 24 [Jatropha curcas] |
| 19 |
Hb_003436_020 |
0.073434643 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 20 |
Hb_016777_040 |
0.0734896473 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |