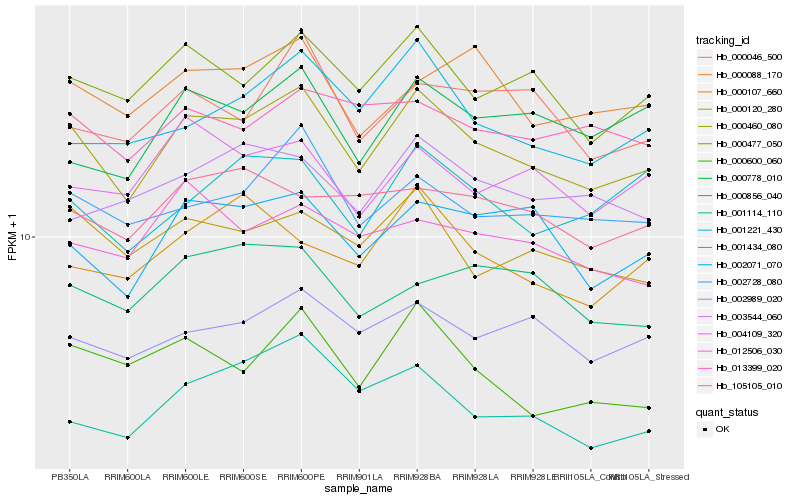
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000460_080 |
0.0 |
- |
- |
acyl-coenzyme A binding domain containing, putative [Ricinus communis] |
| 2 |
Hb_000477_050 |
0.0626733162 |
- |
- |
PREDICTED: 26S protease regulatory subunit 8 homolog A [Jatropha curcas] |
| 3 |
Hb_002071_070 |
0.0678050072 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002728_080 |
0.0678133196 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 5 |
Hb_000120_280 |
0.0692104558 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 6 |
Hb_002989_020 |
0.0715172055 |
- |
- |
PREDICTED: uncharacterized protein LOC105630013 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001434_080 |
0.07537354 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |
| 8 |
Hb_000088_170 |
0.0766631647 |
- |
- |
PREDICTED: ran-binding protein 10-like [Jatropha curcas] |
| 9 |
Hb_001221_430 |
0.0772500799 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At5g63520 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000600_060 |
0.0785064356 |
- |
- |
PREDICTED: uncharacterized protein LOC105633515 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004109_320 |
0.0794297932 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 12 |
Hb_000046_500 |
0.0798525279 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Jatropha curcas] |
| 13 |
Hb_000778_010 |
0.0799617506 |
- |
- |
hypothetical protein [Bacillus subtilis] |
| 14 |
Hb_012506_030 |
0.0809036164 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 15 |
Hb_003544_060 |
0.0809868153 |
- |
- |
hypothetical protein JCGZ_22100 [Jatropha curcas] |
| 16 |
Hb_013399_020 |
0.0812164139 |
- |
- |
PREDICTED: developmentally-regulated G-protein 2 [Jatropha curcas] |
| 17 |
Hb_000856_040 |
0.0815241963 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105640470 [Jatropha curcas] |
| 18 |
Hb_001114_110 |
0.0817558454 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 19 |
Hb_105105_010 |
0.082076489 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 132 [Jatropha curcas] |
| 20 |
Hb_000107_660 |
0.082238837 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |