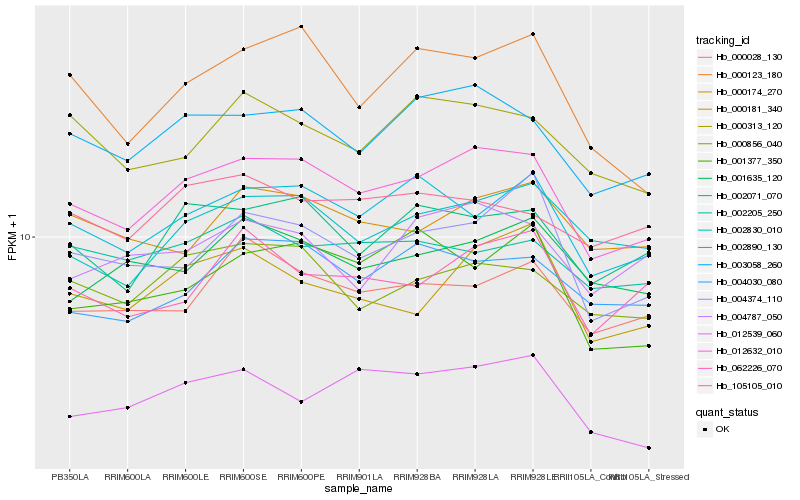
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012632_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640019 [Jatropha curcas] |
| 2 |
Hb_003058_260 |
0.0503362525 |
- |
- |
hypothetical protein POPTR_0003s13040g [Populus trichocarpa] |
| 3 |
Hb_002071_070 |
0.0617122051 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002890_130 |
0.0638531547 |
- |
- |
tip120, putative [Ricinus communis] |
| 5 |
Hb_000856_040 |
0.0649450879 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105640470 [Jatropha curcas] |
| 6 |
Hb_002830_010 |
0.0668932722 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 7 |
Hb_004374_110 |
0.0721140568 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 8 |
Hb_000181_340 |
0.0721507568 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 9 |
Hb_004030_080 |
0.0722346587 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |
| 10 |
Hb_000123_180 |
0.0742561847 |
- |
- |
2-oxoglutarate dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_000028_130 |
0.0742995424 |
- |
- |
PREDICTED: uncharacterized protein LOC105645303 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000313_120 |
0.0756114387 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 13 |
Hb_062226_070 |
0.0769825972 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000174_270 |
0.0770156093 |
- |
- |
PREDICTED: type II inositol 1,4,5-trisphosphate 5-phosphatase FRA3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001635_120 |
0.0774038066 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |
| 16 |
Hb_004787_050 |
0.0783512026 |
- |
- |
PREDICTED: RINT1-like protein MAG2 [Jatropha curcas] |
| 17 |
Hb_012539_060 |
0.0789518872 |
- |
- |
PREDICTED: protein EMBRYONIC FLOWER 1 [Jatropha curcas] |
| 18 |
Hb_002205_250 |
0.0807431742 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_105105_010 |
0.0809622756 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 132 [Jatropha curcas] |
| 20 |
Hb_001377_350 |
0.0817655199 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |