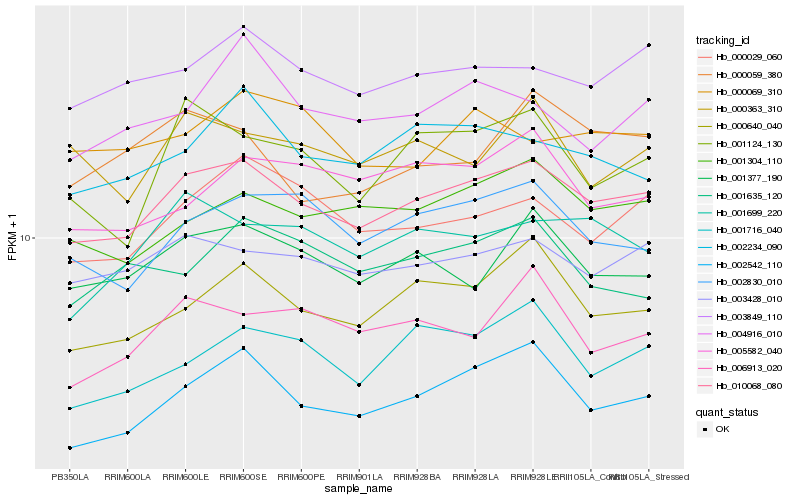
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010068_080 |
0.0 |
- |
- |
PREDICTED: RNA-binding protein rsd1-like [Jatropha curcas] |
| 2 |
Hb_003428_010 |
0.0558585506 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Jatropha curcas] |
| 3 |
Hb_003849_110 |
0.06017059 |
- |
- |
hypothetical protein JCGZ_17842 [Jatropha curcas] |
| 4 |
Hb_001304_110 |
0.0607305491 |
transcription factor |
TF Family: WRKY |
WRKY protein [Hevea brasiliensis] |
| 5 |
Hb_000640_040 |
0.0614863927 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002542_110 |
0.0629749453 |
- |
- |
PREDICTED: uncharacterized protein LOC105642479 [Jatropha curcas] |
| 7 |
Hb_000029_060 |
0.0649341016 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 8 |
Hb_002830_010 |
0.0652890878 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 9 |
Hb_001377_190 |
0.065915888 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002234_090 |
0.0695542651 |
- |
- |
PREDICTED: probable transcriptional regulator SLK2 [Jatropha curcas] |
| 11 |
Hb_001699_220 |
0.0710218866 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH121-like [Jatropha curcas] |
| 12 |
Hb_001124_130 |
0.0727663618 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 13 |
Hb_001716_040 |
0.0731126585 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |
| 14 |
Hb_004916_010 |
0.0758527412 |
- |
- |
PREDICTED: uncharacterized protein LOC105631086 [Jatropha curcas] |
| 15 |
Hb_000363_310 |
0.076662318 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 16 |
Hb_006913_020 |
0.0768665718 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005582_040 |
0.0769880862 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 18 |
Hb_000059_380 |
0.0794796352 |
- |
- |
PREDICTED: uncharacterized protein LOC105648668 [Jatropha curcas] |
| 19 |
Hb_000069_310 |
0.0795861146 |
- |
- |
ubiquitin ligase [Cucumis melo subsp. melo] |
| 20 |
Hb_001635_120 |
0.0796629112 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |