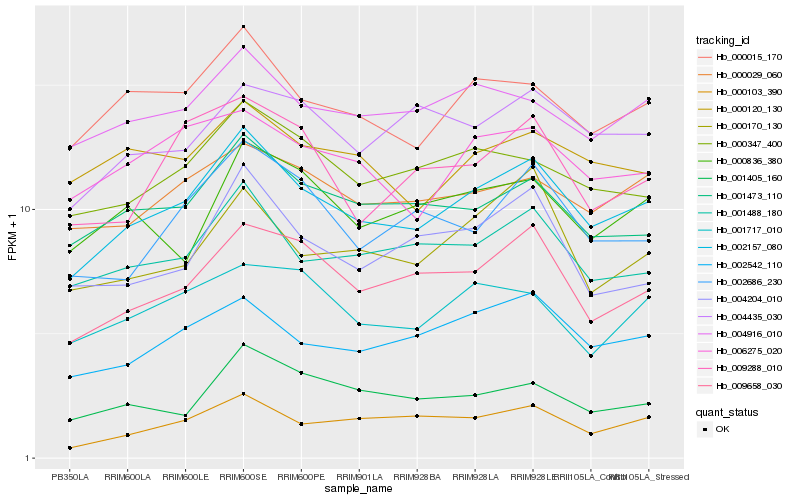
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002157_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000015_170 |
0.0738629663 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002542_110 |
0.0757720917 |
- |
- |
PREDICTED: uncharacterized protein LOC105642479 [Jatropha curcas] |
| 4 |
Hb_001488_180 |
0.0766813892 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 5 |
Hb_009658_030 |
0.0767071131 |
- |
- |
PREDICTED: AP-5 complex subunit beta-1 [Jatropha curcas] |
| 6 |
Hb_001473_110 |
0.0801774881 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C-like [Jatropha curcas] |
| 7 |
Hb_000347_400 |
0.0835279308 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004916_010 |
0.0848882793 |
- |
- |
PREDICTED: uncharacterized protein LOC105631086 [Jatropha curcas] |
| 9 |
Hb_000029_060 |
0.0862961273 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 10 |
Hb_002686_230 |
0.0894819153 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 11 |
Hb_006275_020 |
0.0902307783 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 12 |
Hb_000103_390 |
0.0915179015 |
- |
- |
Pentatricopeptide repeat superfamily protein [Theobroma cacao] |
| 13 |
Hb_004435_030 |
0.0941270579 |
- |
- |
PREDICTED: F-box protein SKIP22 [Jatropha curcas] |
| 14 |
Hb_004204_010 |
0.0947674201 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 15 |
Hb_000170_130 |
0.0948950388 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 16 |
Hb_000120_130 |
0.0957033127 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 17 |
Hb_001405_160 |
0.0959988044 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g09680 [Jatropha curcas] |
| 18 |
Hb_001717_010 |
0.0960480822 |
- |
- |
F-box and wd40 domain protein, putative [Ricinus communis] |
| 19 |
Hb_000836_380 |
0.096974959 |
- |
- |
PREDICTED: C-terminal binding protein AN-like [Populus euphratica] |
| 20 |
Hb_009288_010 |
0.0971955224 |
desease resistance |
Gene Name: Clp_N |
ERD1 protein, chloroplast precursor, putative [Ricinus communis] |