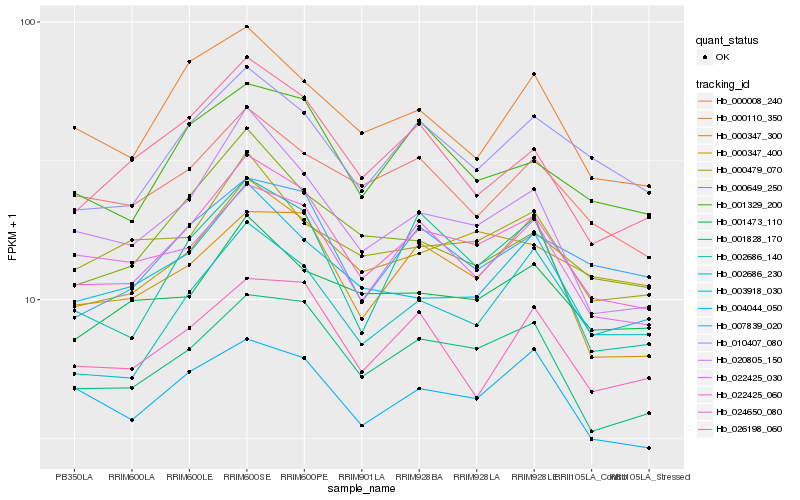
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022425_060 |
0.0 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 2 |
Hb_001828_170 |
0.0499309185 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 3 |
Hb_020805_150 |
0.0617436305 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_001329_200 |
0.0638372144 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 1 [Jatropha curcas] |
| 5 |
Hb_010407_080 |
0.0669051519 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007839_020 |
0.0707715219 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003918_030 |
0.0709032877 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 1C-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000347_400 |
0.071207591 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_022425_030 |
0.0713017243 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 10 |
Hb_000347_300 |
0.0722418518 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 11 |
Hb_000008_240 |
0.0728358922 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000479_070 |
0.0728492278 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 13 |
Hb_026198_060 |
0.0731922931 |
- |
- |
PREDICTED: calcium-dependent protein kinase 11 [Jatropha curcas] |
| 14 |
Hb_004044_050 |
0.0737246995 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 15 |
Hb_000649_250 |
0.0747899 |
transcription factor |
TF Family: C2H2 |
PREDICTED: UBX domain-containing protein 6 [Jatropha curcas] |
| 16 |
Hb_000110_350 |
0.0760048053 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 17 |
Hb_001473_110 |
0.0770497369 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C-like [Jatropha curcas] |
| 18 |
Hb_002686_230 |
0.0775654004 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_002686_140 |
0.0776039794 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_024650_080 |
0.0788988608 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g09030 [Jatropha curcas] |