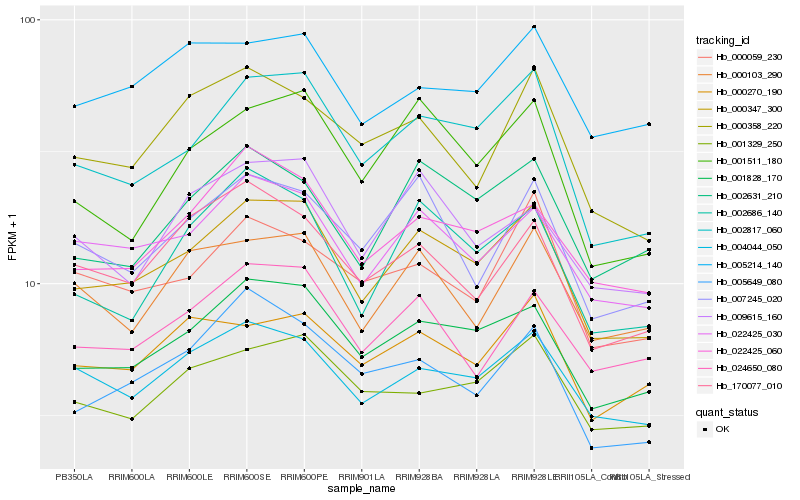
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_300 |
0.0 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 2 |
Hb_001828_170 |
0.0472720921 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 3 |
Hb_002817_060 |
0.0483287169 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 4 |
Hb_022425_030 |
0.0610127148 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 5 |
Hb_004044_050 |
0.0684650912 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 6 |
Hb_022425_060 |
0.0722418518 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 7 |
Hb_002686_140 |
0.074882169 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001511_180 |
0.0751133588 |
- |
- |
Clathrin heavy chain 2 -like protein [Gossypium arboreum] |
| 9 |
Hb_002631_210 |
0.0772768941 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 10 |
Hb_001329_250 |
0.0784747662 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 11 |
Hb_007245_020 |
0.0798479053 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 12 |
Hb_024650_080 |
0.0801922064 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g09030 [Jatropha curcas] |
| 13 |
Hb_000270_190 |
0.0817601719 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |
| 14 |
Hb_000103_290 |
0.0819996796 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 15 |
Hb_000358_220 |
0.0834440658 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 16 |
Hb_005649_080 |
0.0835169946 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 17 |
Hb_170077_010 |
0.0837527948 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 18 |
Hb_009615_160 |
0.0846164733 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 19 |
Hb_000059_230 |
0.0847427457 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 20 |
Hb_005214_140 |
0.0849385883 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |