| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
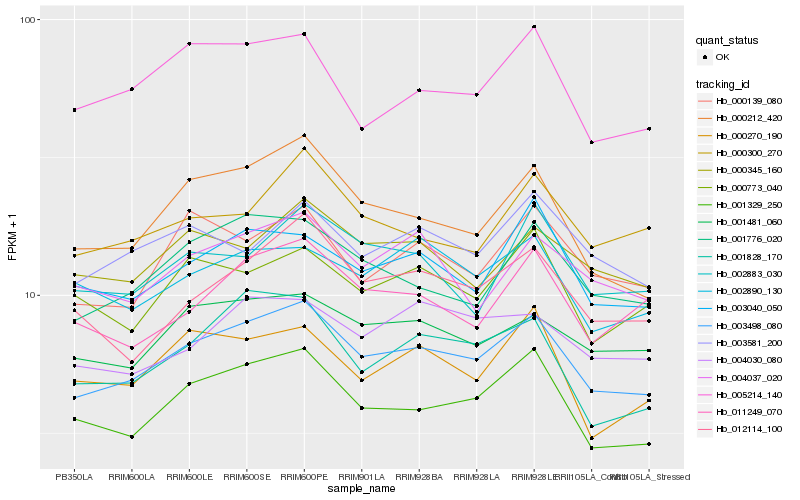
Hb_003498_080 |
0.0 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 2 |
Hb_001329_250 |
0.0517195621 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000212_420 |
0.0598998846 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 4 |
Hb_001776_020 |
0.0600112955 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 5 |
Hb_001481_060 |
0.0626648202 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003040_050 |
0.0643023601 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 7 |
Hb_012114_100 |
0.0686501872 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 8 |
Hb_005214_140 |
0.0692645518 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 9 |
Hb_000139_080 |
0.0693344103 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001828_170 |
0.069939543 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 11 |
Hb_004037_020 |
0.0700190833 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000345_160 |
0.070238515 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 13 |
Hb_011249_070 |
0.0719990355 |
- |
- |
hypothetical protein POPTR_0018s12610g [Populus trichocarpa] |
| 14 |
Hb_003581_200 |
0.0730917263 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |
| 15 |
Hb_000773_040 |
0.0736251039 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000300_270 |
0.0737007857 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 17 |
Hb_004030_080 |
0.0748156735 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |
| 18 |
Hb_002883_030 |
0.0748290548 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 19 |
Hb_002890_130 |
0.0750297922 |
- |
- |
tip120, putative [Ricinus communis] |
| 20 |
Hb_000270_190 |
0.0752312437 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |