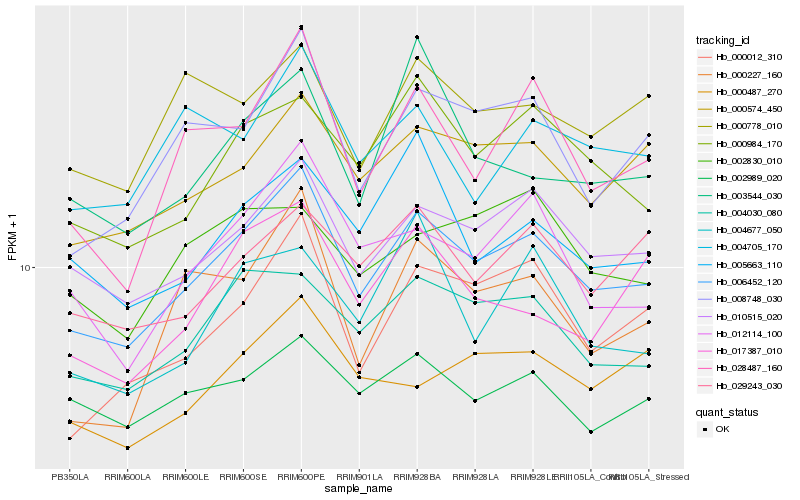
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006452_120 |
0.0 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 2 |
Hb_010515_020 |
0.0457268037 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 3 |
Hb_004030_080 |
0.0564514715 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |
| 4 |
Hb_000574_450 |
0.0565369433 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein 2 [Jatropha curcas] |
| 5 |
Hb_008748_030 |
0.0595887546 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000984_170 |
0.0601907293 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 7 |
Hb_000012_310 |
0.062401103 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_012114_100 |
0.0624114824 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 9 |
Hb_028487_160 |
0.0638740637 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 10 |
Hb_029243_030 |
0.0684139473 |
- |
- |
PREDICTED: regulatory-associated protein of TOR 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005663_110 |
0.0695656345 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002989_020 |
0.0708275949 |
- |
- |
PREDICTED: uncharacterized protein LOC105630013 isoform X2 [Jatropha curcas] |
| 13 |
Hb_017387_010 |
0.0709356303 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 14 |
Hb_000778_010 |
0.0712434573 |
- |
- |
hypothetical protein [Bacillus subtilis] |
| 15 |
Hb_000227_160 |
0.0727669247 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 16 |
Hb_000487_270 |
0.0729115679 |
- |
- |
hypothetical protein JCGZ_22651 [Jatropha curcas] |
| 17 |
Hb_004705_170 |
0.0730077886 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 18 |
Hb_003544_030 |
0.0742673529 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 19 |
Hb_004677_050 |
0.0743207922 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002830_010 |
0.0744225186 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |